Figure 6.
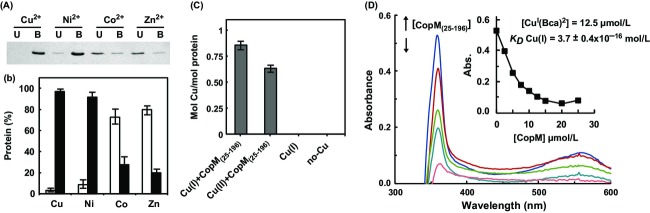
CopM binds copper in vitro. (A) Analysis of CopM(25-196) protein interaction with metals. His-Bind resin columns were loaded with 5 mmol/L CuSO4, NiSO4, ZnSO4, and CoCl2. Hundred micrograms purified CopM(25-196) protein was applied to the columns. The unbound (U lanes) and bound (B lanes) fractions were analyzed by 15% SDS-PAGE and Coomassie blue staining. (B) Quantification of CopM(25-196) in bound and unbound fractions. Coomassie-stained gel was scanned and bands intensity was quantified using ImageJ program; the graph represents the average of three independent experiments. Unbound fraction (white), bound fraction (black). (C) Specificity of copper ion binding by CopM(25-196). CopM of 800 μg was incubated for 10 min with 1 mmol/L Cu(I) or Cu(II) and gel filtration was used to remove unbound Cu ions, which were measured by titration with BCS as described in “Experimental Procedures” section. Bar graph represents mol of Cu/mol of CopM from three independent experiments. Error bars represent SE. Cu(I), only copper; no-Cu only CopM(25-196). (D) Determination of the Cu(I) dissociation constant, KD, of CopM(25-196) by titration into a solution of 60 μmol/L BCA. The graph shows the spectral changes of the [Cu(BCA)2]3− form on the CopM(25-196) titration. BCA (60 μmol/L) in 20 mmol/L Tris-HCl (pH 7.5), 50 mmol/L NaCl was titrated against CopM (0–25 μmol/L) measuring absorbance in the 350–600 nm range. Inset: The decrease at 358 nm relative to CopM(25-196) additions for a [Cu(BCA)2]FINAL = 1.4 μmol/L. Dissociation constant, KD, was estimated as described in “Experimental Procedures” section from three independent experiments like the one shown in D.
