Abstract
The tongue is a muscular organ that is essential in vertebrates for important functions, such as food intake and communication. Little is known about regulation of myogenic progenitors during tongue development when compared with the limb or trunk region. In this study, we investigated the relationship between different myogenic subpopulations and the function of canonical Wnt signaling in regulating these subpopulations. We found that Myf5- and MyoD-expressing myogenic subpopulations exist during embryonic tongue myogenesis. In the Myf5-expressing myogenic progenitors, there is a cell-autonomous requirement for canonical Wnt signaling for cell migration and differentiation. In contrast, the MyoD-expressing subpopulation does not require canonical Wnt signaling during tongue myogenesis. Taken together, our results demonstrate that canonical Wnt signaling differentially regulates the Myf5- and MyoD-expressing subpopulations during tongue myogenesis.
Keywords: craniofacial muscles, myogenesis, progenitors, Myf5, MyoD, Wnt/β-catenin
Introduction
The tongue emerged as an essential organ during vertebrate evolution due to its role in food intake. Moreover, in humans and other species, it exerts important functions in speech and communication (Parada et al. 2012). The appropriate prenatal development of tongue skeletal muscle is essential for these physiologic functions. During development, extrinsic signals and intrinsic regulating factors guide myogenic progenitors to self-renew, differentiate, and eventually form the mature tongue musculature. Tongue myogenic progenitors originate from the anterior-most somites and migrate through bilateral pathways that together form the hypoglossal cord and, ultimately, the tongue primordium (Sambasivan et al. 2011). After migration into the craniofacial region, the tongue myogenic progenitors initiate intimate interactions with cranial neural crest cells (Hosokawa et al. 2010). Our previous studies have demonstrated that there is a cell-autonomous requirement for Smad4-mediated TGFβ signaling during myogenic differentiation and myoblast fusion in the tongue (Han et al. 2012). Yet, to date, the different signals involved and the mechanisms by which tissue-tissue interactions precisely regulate this delicate developmental process remain unclear.
Skeletal muscle development takes place in a succession of overlapping steps termed embryonic myogenesis and fetal myogenesis (Biressi et al. 2007). During embryonic myogenesis, myogenic progenitors express a number of myogenic regulatory factors, including Myf5 and MyoD (Kablar et al. 1997; Braun and Gautel 2011). Mice lacking both are virtually devoid of myoblasts and myofibers (Rudnicki et al. 1993). Myf5 is the earliest myogenic regulatory factor involved in myoblast specification and maintenance. It is expressed in somitic and cranial mesoderm and is detectable as early as E8 (Haldar et al. 2007). Myf5 expression subsequently decreases and is undetectable by E14 (Ott et al. 1991). MyoD expression begins later than that of Myf5 at E10.5 in the somites and limb buds, and high levels of MyoD are maintained throughout muscle development (Sassoon et al. 1989; Yamamoto et al. 2009). In the limbs, there is considerable heterogeneity in MyoD and Myf5 protein expression pattern from E11.5 to E12.5 (Haldar et al. 2008). DTA-mediated cell population ablation experiments have suggested that there are Myf5-dependent and Myf5-independent myogenic lineages during embryonic myogenesis. MyoD-expressing cells can compensate for the loss of the Myf5-dependent myogenic lineage after ablation (Gensch et al. 2008). A recent study has reported that the majority of Myf5(+) progenitors eventually become MyoD(+) progenitors in the limbs (Wood et al. 2013). To date, little is known about the relationship between MyoD(+) and Myf5(+) myogenic populations, although this relationship is important for understanding compensatory mechanisms during skeletal muscle development.
The canonical Wnt cascade has emerged as a critical regulator of stem cells and progenitors involved in development and tissue regeneration (Reya and Clevers 2005). During skeletal muscle development, canonical Wnt signaling plays important roles in dermomyotome and myotome formation (Ikeya and Takada 1998; Linker et al. 2003; Parker et al. 2003; Otto et al. 2006). In the somites, Wnt1 ligands preferentially activate Myf5, whereas Wnt7a ligands activate MyoD in myogenic progenitors (Tajbakhsh et al. 1998). It was recently reported that canonical Wnt signaling exerts different functions during embryonic and fetal myogenesis (Hutcheson et al. 2009). During embryonic myogenesis, canonical Wnt signaling is essential for the delamination of the Pax3-dependent myogenic population in the limb. During fetal myogenesis, canonical Wnt signaling is critical for the formation of myofibers with the correct number and type in the Pax7-dependent myogenic population.
In this study, we further investigated the functional significance of canonical Wnt signaling in regulating Myf5- and MyoD-expressing progenitors during tongue development. We conclude that Myf5(+) progenitors and MyoD(+) progenitors are different myogenic subpopulations and that canonical Wnt signaling regulates these 2 subpopulations differentially. This is an important discovery through which we will have a better understanding of the molecular and cellular regulatory mechanisms of tongue myogenesis.
Materials and Methods
Mice
Myf5-Cre (Tallquist et al. 2000), MyoD-Cre (Yamamoto et al. 2009), ROSA26LoxP-STOP-LoxPLacZ (Soriano, 1999), ROSA26LoxP-STOP-LoxP-Tdtomato (Madisen et al. 2010), ROSA26LoxP-STOP-LoxP-DTA (Voehringer et al. 2008), Ctnnb1flox/flox (Brault et al. 2001), and Axin2-LacZ (Lustig et al. 2002) mice have been described. Genotyping was carried out through polymerase chain reaction on tail tip or yolk sac DNA.
Histologic Analysis
All samples were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS; 20mM sodium phosphate, 0.10M NaCl, pH 7.4), processed through serially increasing dilutions of ethanol, paraffin embedded, and sectioned through routine procedures. For general morphology, deparaffinized sections were stained with hematoxylin and eosin via standard procedures.
β-galactosidase Activity Assays
E10.5 embryos were harvested and stained for β-galactosidase (β-gal) activity according to standard procedures (Chai et al. 2000). For detection of β-gal activity, samples from mice were fixed in 0.2% glutaraldehyde and decalcified with 10% ethylenediaminetetraacetic acid for 2 wk. We cut frozen sections of 10-µm thickness and analyzed β-gal activity following standard protocols.
Immunofluorescence
Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C, immersed in 15% sucrose in PBS overnight at 4°C, followed by 30% sucrose overnight, then immersed in OCT compound (Sakura Finetek) and frozen on dry ice. Embryos were stored at −80°C.
Frozen embryos were cryostat sectioned into 10×µm slices, collected on Superfrost Plus slides (Fisher), and stored at −20°C. Antigen retrieval was used for detection of MyHC. For antigen retrieval, slides were washed 3 times for 5 min each in PBS, then placed in 0.1M Tris buffer at pH 9.0 and incubated in a 1200W microwave at 10% power for 30 min, followed immediately by incubation in blocking buffer.
The following antibodies and working dilutions were used for immunofluorescence.
MyoD: monoclonal antibody 5.8A (1:100 dilution; catalog M351201-2, Dako)
Myf5: rabbit polyclonal antibody (1:100 dilution; catalog sc-302, Santa Cruz Biotechnology)
MyHC: monoclonal antibody MF20 (1:100 dilution; Developmental Studies Hybridoma Bank)
β-gal: chicken polyclonal antibody (1:50 dilution, catalog ab9361, Abcam), Alexa Fluor 568 goat anti-mouse IgG (1:200 dilution, catalog A-11004, Invitrogen), Alexa Fluor 488 goat anti-mouse IgG (1:200 dilution, catalog A-11001, Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (1:200 dilution, catalog A-11008, Invitrogen), Alexa Fluor 568 goat anti-chicken IgG (1:200 dilution; catalog A-11041, Invitrogen)
For detection of MyHC, sections were blocked in TNB blocking buffer (0.5% blocking reagent, catalog FP1020, PerkinElmer; 0.1M Tris-HCl, pH 7.5; 0.15M NaCl) for 1 h at room temperature after antigen retrieval, washed 2 times in TNT wash buffer (0.1M Tris-HCl, pH 7.5; 0.15M NaCl; 0.05% Tween20), and incubated in primary antibody suspended in TNB blocking buffer overnight at 4°C. After three 10-min washes in TNT wash buffer, sections were incubated for 1 h in 1:200 dilution of Alexa Fluor 568 goat anti-mouse IgG. After three 10-min washes in TNT wash buffer, sections were counterstained with DAPI (0.1 µg/mL), and slides were coverslipped with Fluoro-Gel (catalog 17985-11, Electron Microscopy Sciences).
Simultaneous detection of MyoD/Myf5 and MyoD/β-gal followed the MyHC protocol without antigen retrieval. Sections were incubated with both primary antibodies simultaneously, washed in TNT wash buffer, and incubated with both secondary antibodies simultaneously.
Tongue Area Measurement
For quantification of tongue area measurement, hematoxylin and eosin staining sections were prepared from 3 sets of E18.5 Myf5-Cre;β-cateninfl/+ control and Myf5-Cre;β-cateninfl/fl mouse embryos as well as 3 sets of newborn MyoD-Cre;β-cateninfl/+ control and MyoD-Cre;β-cateninfl/fl mouse embryos, and 3 sections were scored per sample. The same procedure was used for Myf5-Cre;R26RDTA/+ and control, as well as MyoD-Cre;R26RDTA/+ and control, mouse embryos at newborn stage for tongue area measurement. All measurements and cell counting were performed with Image-J 1.46r software. Results were assessed for statistical significance with Student’s t tests.
Statistics
SPSS 13.0 was used to perform statistical analysis. Significance was assessed by independent 2-tailed Student’s t tests; P values < 0.05 were considered significant.
Results
Differences between Myf5-expressing and MyoD-expressing Subpopulations during Tongue Myogenesis
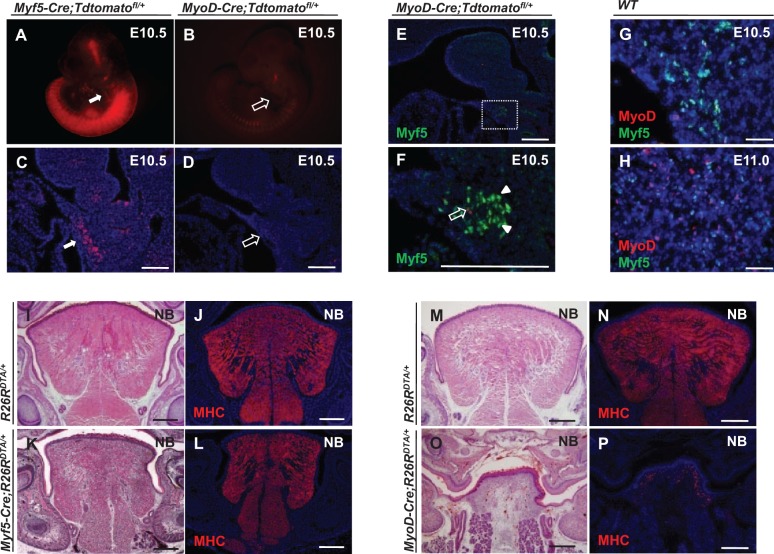
We analyzed the expression patterns of Myf5 and MyoD using Myf5-Cre;Tdtomatofl/+ and MyoD-Cre;Tdtomatofl/+ reporter lines. Myf5-derived myogenic progenitors are already present in the hypoglossal cord at E10.5, whereas only a few MyoD-derived myogenic progenitors have migrated into the hypoglossal cord (Fig. 1A–D). When we assayed the Myf5 protein expression pattern in the hypoglossal cord in MyoD-Cre;Tdtomatofl/+ mice at E10.5, we found that cells positive for Myf5 are not derived from MyoD-expressing cells (Fig. 1E, F). We also used coimmunostaining of Myf5 and MyoD in wild-type mice to confirm these results. We found that only a few coexpressing cells are detectable at E10.5 in the hypoglossal cord. At E11.0, only a small number of myogenic progenitors coexpress both Myf5 and MyoD in the tongue bud (Fig. 1G, H).
Figure 1.
Myf5- and MyoD-expressing cells are distinct subpopulations during tongue myogenesis. (A–D) Detection of Myf5- and MyoD-derived cells in E10.5 Myf5-Cre;Tdtomatofl/+ (A, C) and MyoD-Cre;Tdtomatofl/+ (B, D) reporter mice, respectively. Myf5-derived cells are detectable in the hypoglossal cord (white arrows), whereas few MyoD-derived cells are detectable (open arrows). (E, F) Myf5 immunostaining (green, arrowheads) in the hypoglossal cord region of E10.5 MyoD-Cre;Tdtomatofl/+ reporter mice (MyoD-derived myogenic cells indicated by red, open arrow). Boxed area (E) is shown magnified (F). (G, H) Double immunostaining of MyoD (red) and Myf5 (green) in the hypoglossal cord at E10.5 (G) and the tongue bud at E11.0 (H) of wild-type mice. (I–L) Hematoxylin and eosin staining (I, K) and myosin heavy chain (MHC) immunostaining (J, L) of tongues from newborn (NB) Myf5-Cre;R26RDTA/+ and R26RDTA/+ control mice. (M–P) Hematoxylin and eosin staining (M, O) and MHC immunostaining (N, P) of tongues from NB MyoD-Cre;R26RDTA/+ and R26RDTA/+ control mice. Scale bars, 200 µm.
Next, we crossed Myf5-Cre mice with R26RDTA/+ mice to ablate the Myf5-expressing progenitors and found that the muscle pattern and size of newborn tongues were only mildly altered when compared with controls (Fig. 1I–L, Appendix Fig. E). This finding is consistent with previous studies demonstrating that the Myf5-independent myogenic lineage could compensate for loss of Myf5-expressing myogenic cells (Gensch et al. 2008; Haldar et al. 2008). Using the same DTA strategy to ablate the MyoD-expressing progenitors, we found that the size of the tongue was much smaller in newborn MyoD-Cre;R26RDTA/+ mice when compared with controls (Fig. 1M, O; Appendix Fig. F). Moreover, we detected a striking reduction in muscle fiber formation in the tongue (Fig. 1N, P). Taken together, our results suggest that the MyoD-expressing, but not the Myf5-expressing, subpopulation is essential for tongue myogenesis.
Activation of Canonical Wnt Signaling in Myogenic Progenitors during Tongue Myogenesis
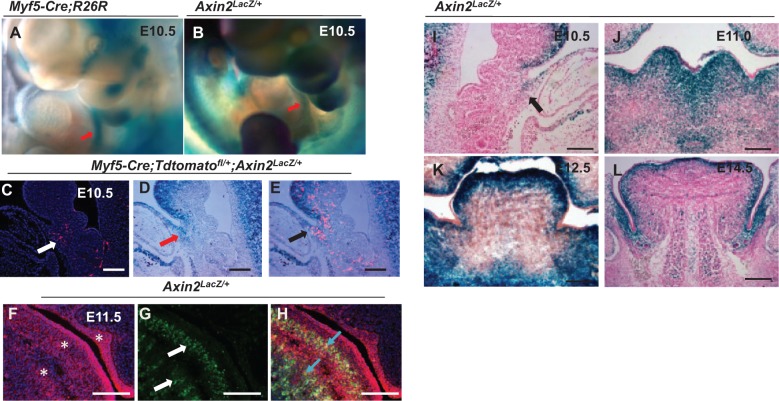
Our current study indicated that canonical Wnt signaling is activated in the myogenic region during tongue embryonic myogenesis. We utilized the Axin2Lacz/+ reporter line, which serves as a readout of canonical Wnt signaling, to detect activation of the Wnt signaling cascade during tongue myogenesis. We found that canonical Wnt signaling is activated in the same region of the hypoglossal cord as the Myf5-derived population (Fig. 2A, B). We confirmed this finding using Myf5-Cre;Tdtomatofl/+;Axin2LacZ/+ mice to compare LacZ staining and Myf5-derived lineage labeling in adjacent sections (Fig. 2C–E). We also examined Wnt activity in the MyoD+ myogenic population using E11.5 Axin2lacZ+/- mouse embryos and found that Wnt signaling is activated in the MyoD+ population at E11.5 (Figure 2F–H). Next, we analyzed canonical Wnt signaling activity in the tongue at embryonic stages from E10.5 to E14.5. Canonical Wnt signaling is activated in the myogenic region of the tongue at E10.5 (Fig. 2I), then gradually decreases in the later embryonic stages. In later stages of tongue development, activated Wnt signaling shifts from the myogenic region into the surrounding cranial neural crest–derived supportive tissue (Fig. 2J–L).
Figure 2.
Activation of canonical Wnt signaling in myogenic progenitors during tongue myogenesis. (A) Whole-mount X-gal staining of E10.5 Myf5-Cre;R26R embryos. Red arrow indicates Myf5-derived cells in the hypoglossal cord. (B) Whole-mount X-gal staining of E10.5 Axin2LacZ/+ embryos. Red arrow indicates activation of the canonical Wnt signaling pathway in the hypoglossal cord region. (C–E) Visualization of Myf5-derived cells (C; red) and lacZ staining indicating activation of canonical Wnt signaling (D; blue) in adjacent sections of Myf5-Cre;Tdtomatofl/+;Axin2LacZ/+ mice. White arrow indicates Myf5 derived cells. Red arrow indicates Axin2+ cells. Colocalization is indicated by the black arrow (E). (F–H) Immunohistochemical staining of β-galactosidase (F; asterisks) and MyoD (G; white arrows) in E11.5 Axin2LacZ/+ mice embryos. Blue arrows (H) indicate colocalization. (I–L) X-gal staining of tongue buds in Axin2LacZ/+ mice at E10.5 (I), E11.5 (J), E12.5 (K), and E14.5 (L) shows activation of canonical Wnt signaling pathway. Black arrow indicates the hypoglossal cord (I). Scale bars, 200 µm.
Loss of β-catenin Leads to Migration and Differentiation Defects in the Myf5-dependent Subpopulation
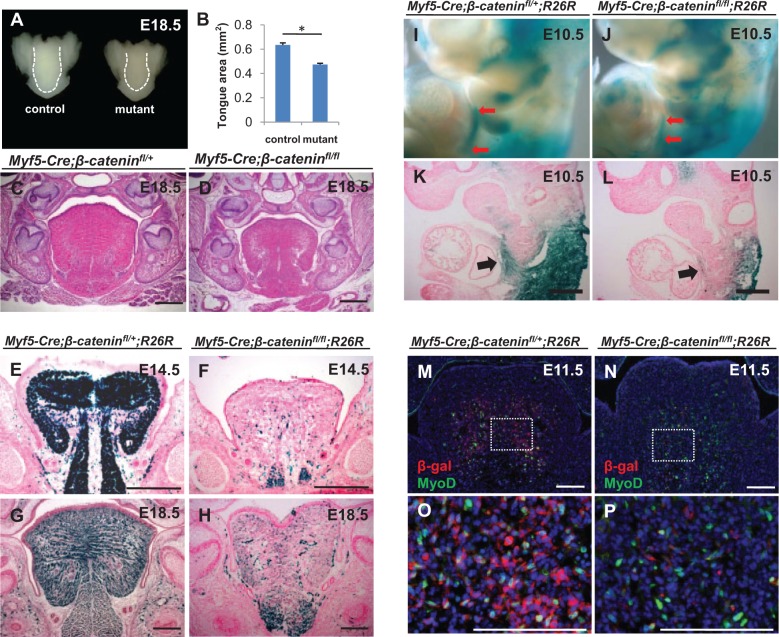
To test the functional significance of canonical Wnt signaling in regulating tongue myogenesis, we generated Myf5-Cre;β-cateninfl/fl mice and found that the size of the tongue is smaller at birth after loss of β-catenin in the Myf5-expressing subpopulation (Fig. 3A–D). Lineage tracing of the Myf5-derived muscle fibers in Myf5-Cre;β-cateninfl/fl;R26R mice from E11.5 to E18.5 demonstrated that only a few of the muscle fibers are derived from Myf5-expressing cells, suggesting that most muscle fibers are derived from Myf5-independent myogenic cells (Fig. 3E–H, Appendix Fig. A–D). After loss of β-catenin, the length of the Myf5-derived hypoglossal cord was reduced in E10.5 mutant embryos when compared with controls, as assessed by whole-mount LacZ staining (Fig. 3I, J). Also in LacZ-stained sections, the number of Myf5-derived cells in the hypoglossal cord was reduced at E10.5 when compared with controls. A large number of Myf5-derived myogenic cells failed to migrate into the tongue bud (Fig. 3K, L). The percentage of Myf5-derived cells expressing MyoD was significantly reduced in Myf5-Cre;β-cateninfl/fl;R26R mice as compared with controls (Fig. 3M–P, Appendix Fig. G). Taken together, our results suggest that myogenic cell migration and differentiation are both compromised after loss of β-catenin in the Myf5-expressing subpopulation during embryonic tongue myogenesis.
Figure 3.
Loss of β-catenin leads to migration and differentiation defects in the Myf5-expressing subpopulation. (A–D) Whole tongues (A) and hematoxylin and eosin staining of tongue sections (C, D) from E18.5 Myf5-Cre;β-cateninfl/fl (mutant) and Myf5-Cre; β-cateninfl/+ (control) mice. Quantification of the tongue area confirmed a significant difference in tongue size between Myf5-Cre;β-cateninfl/fl and control mice at E18.5 (B). *P < 0.05, n = 3. (E–H) X-gal staining of sections of tongues from E14.5 (E, F) and E18.5 (G, H) Myf5-Cre;β-cateninfl/fl;R26R and Myf5-Cre;β-cateninfl/+;R26R control mice. (I–L) Whole-mount X-gal staining (I, J) and X-gal staining of sections (K, L) of E10.5 Myf5-Cre;β-cateninfl/fl;R26R and Myf5-Cre;β-cateninfl/+;R26R control mice. Red arrows indicate migration defects of Myf5-derived cells in the hypoglossal cord (I, J). Black arrows highlight migration defects in sections from Myf5-Cre;β-cateninfl/fl; R26R mice (K, L). (M–P) Double immunostaining of MyoD (green) and β-gal (red) in E11.5 Myf5-Cre;β-cateninfl/fl;R26R and Myf5-Cre;β-cateninfl/+;R26R control mice. Boxed areas (M, N) are shown magnified (O, P). Scale bars, 200 µm.
Canonical Wnt Signaling Is Not Required in the MyoD-expressing Subpopulation
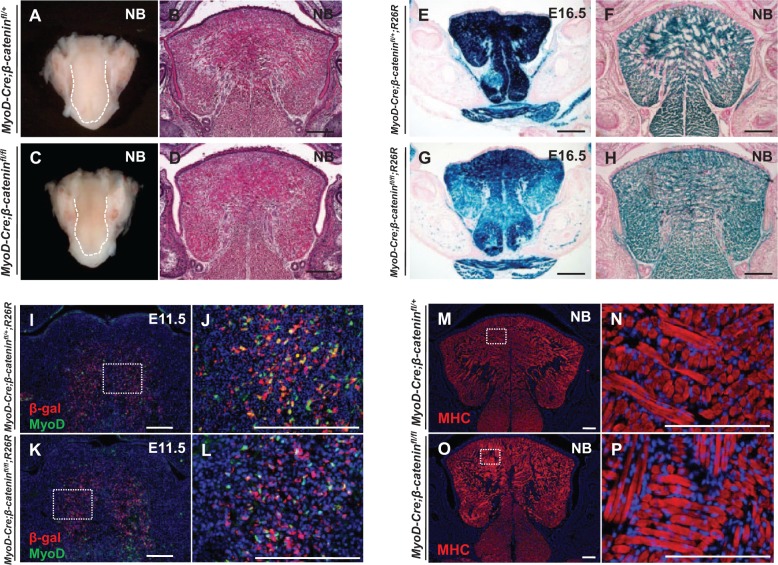
In parallel, we also generated MyoD-Cre;β-cateninfl/fl mice and found that the size and morphology of tongue were not affected after deletion of β-catenin in MyoD-expressing cells (Fig. 4A–D, Appendix Fig. H). Lineage tracing of MyoD-derived muscle fibers in E16.5 and newborn MyoD-Cre;β-cateninfl/fl;R26R mice indicated that almost all the muscle fibers in the tongue were derived from the MyoD-expressing subpopulation and no significant tongue muscle pattern defects were detectable (Fig. 4E–H). We analyzed MyoD expression in the MyoD-derived myogenic cells after deletion of β-catenin in MyoD-Cre;β-cateninfl/fl;R26R mice and found that it was indistinguishable from controls (Fig. 4I–L, Appendix Fig. I). We also assessed the expression of myosin heavy chain, a marker for mature myofibers, to determine whether the differentiation of tongue muscle was affected by loss of β-catenin in the MyoD-expressing subpopulation. We found that the expression of MHC in MyoD-Cre;β-cateninfl/fl mice was unaffected relative to controls (Fig. 4M–P). Our results therefore suggest that, in contrast to the Myf5-expressing subpopulation, the MyoD-expressing subpopulation does not require canonical Wnt signaling during tongue myogenesis.
Figure 4.
Canonical Wnt signaling is not required in the MyoD-expressing subpopulation. (A–D) Whole tongues (A, C) and hematoxylin and eosin staining (B, D) of tongue sections from newborn (NB) MyoD-Cre;β-cateninfl/fl and MyoD-Cre;β-cateninfl/+ control mice. (E–H) X-gal staining of E16.5 (E, G) and NB (F, H) MyoD-Cre;β-cateninfl/fl;R26R and MyoD-Cre;β-cateninfl/+;R26R control mice. (I–L) Double immunostaining of MyoD (green) and β-gal (red) in E11.5 MyoD-Cre;β-cateninfl/fl;R26R and MyoD-Cre;β-cateninfl/+;R26R control mice. Boxed areas (I, K) are shown magnified (J, L). (M–P) Myosin heavy chain (MHC) immunostaining of NB MyoD-Cre;β-cateninfl/fl and MyoD-Cre;β-cateninfl/+ control mice. Boxed areas (M, O) are shown magnified (N, P). Scale bars, 200 µm.
Discussion
Different Myogenic Subpopulations during Tongue Myogenesis
There is substantial spatiotemporal heterogeneity during skeletal muscle development (Biressi et al. 2007). For example, previous studies based on in situ hybridization and immunostaining in the limb and trunk regions have demonstrated considerable cellular heterogeneity of Myf5(+) and MyoD(+) progenitors during myogenesis. Cells that are positive for either Myf5 or MyoD are far more prevalent than those coexpressing the 2 markers. However, these results provide only a snapshot of myogenic regulatory factor expression at discrete development stages. Cell population–specific ablation mediated by Cre-activated DTA expression is a more powerful means by which to interpret different myogenic subpopulation dynamics. In this study, we found that the hypoglossal cord is almost entirely composed of Myf5-derived myogenic progenitors at E10.5 and that these myogenic progenitors are not derived from MyoD-expressing cells. Also we found that the majority of myogenic progenitors express either Myf5 or MyoD in the hypoglossal cord and the tongue bud from E10.5 to E11.0. Moreover, we used DTA-mediated cell population–specific ablation experiments to detect Myf5- and MyoD-expressing subpopulation dynamics during tongue myogenesis. By ablating the DTA-mediated Myf5-expressing subpopulation, we found that the Myf5-independent lineages can compensate for Myf5-expressing myogenic cell functions during the formation of the skeletal muscular system in the tongue. Consistent with the phenotype previously observed in the limbs, embryos with an ablated Myf5-expressing subpopulation demonstrate almost full recovery of skeletal muscle development in the tongue (Haldar et al. 2008). In contrast, deletion of the MyoD-expressing subpopulation resulted in the failure of tongue skeletal muscle formation. Taken together, these results support the conclusions that Myf5- and MyoD-expressing cells are different subpopulations during early embryonic tongue myogenesis and that other myogenic subpopulations possess a substantial capacity to compensate for the loss of the Myf5-expressing subpopulation in developing tongue skeletal muscle.
We used the tongue as a model to investigate the relationship between the Myf5- and MyoD-expressing subpopulations in the craniofacial region because the developmental mechanisms underlying craniofacial muscle are poorly understood when compared with those of the trunk and limb muscles (Sambasivan et al. 2011; Buckingham and Rigby, 2014). The tongue has unique features due to its location in the transition zone between the trunk and head. Trunk and tongue skeletal muscles share traits because they both originate from somites. In our study, we found that the MyoD-expressing subpopulation could compensate for loss of the Myf5-expressing subpopulation during myogenesis in the tongue, as it can in the limb region but not vice versa. However, the tongue also has traits that differ from trunk muscles because the cranial neural crest cells contribute to connective tissues in the tongue and influence the morphologic patterning of these muscles.
In the limb, MyoD- and Myf5-expressing myogenic cells both emerge around E10.5 (Mankoo et al. 1999; Schuster-Gossler et al. 2007). In contrast, we found that the hypoglossal cord is almost entirely composed of Myf5-expressing cells, whereas few MyoD-derived progenitors are present in the hypoglossal cord at E10.5. One possible explanation for this difference is that the migrating speed of Myf5- and MyoD-expressing subpopulations may vary due to different responses to signals secreted by surrounding cranial neural crest cells.
Wnt Signaling during Myogenic Development
Previous studies have reported that different myogenic progenitors in the same environment may respond differently to the same signals. For instance, TGFβ and/or BMP may inhibit the differentiation of fetal myoblasts and satellite cells but not embryonic myoblasts (Cusella-De Angelis et al. 1994). Different Wnts activate Myf5 and MyoD in somites. Specifically, Myf5 is activated by Wnt1, which functions through a β-catenin-dependent pathway, whereas MyoD is activated by Wnt7 via a β-catenin-independent pathway (Tajbakhsh et al. 1998). Furthermore, canonical Wnt signaling regulates Pax3- and Pax7-dependent myogenic progenitors differentially in the limb region during embryonic and fetal myogenesis (Hutcheson et al. 2009).
Here, we demonstrated that canonical Wnt signaling regulates the migration and differentiation of the Myf5-expressing subpopulation. The compromised migration in the β-catenin knockout mutants might be due to cytoskeletal defects because β-catenin is an essential component of the cytoskeleton. In contrast, the MyoD-expressing subpopulation appears not to require canonical Wnt signaling during tongue myogenesis. An attractive hypothesis is that a degree of functional redundancy exists such that the ability to develop skeletal muscle is maintained by the MyoD-expressing subpopulation even when canonical Wnt signaling has been compromised in the Myf5-expressing subpopulation.
The substantial spatiotemporal heterogeneity during skeletal muscle development may be controlled by different signaling pathways. Previous studies have reported that the Myf5-dependent lineage gives rise to about 50% of adult myonuclei (Haldar et al. 2008). After loss of Myf5-expressing cells, the MyoD-expressing cells expand to compensate for the loss of Myf5 lineage to restore myogenesis. This finding implies that the Myf5(+) and MyoD(+) progenitors have the same and/or overlapping potential to expand and differentiate into mature myofibers.
In the intestinal crypt, the intestinal stem cells double their numbers each day and stochastically adopt stem or transit-amplifying cell fates to determine the expansion or restriction of different subpopulations (Snippert et al. 2010). In the case of muscle stem cell self-renewal during muscle development, Myf5 and MyoD are induced by different Wnt ligands. The question remains as to how the myogenic program is regulated to expand or restrict Myf5(+) and MyoD(+) progenitors during myogenesis. Based on our study, canonical Wnt signaling is a key factor determining Myf5-expressing cell migration and differentiation. Therefore, canonical Wnt signaling controls Myf5(+) progenitors, yet another signaling pathway (or pathways) is required to regulate the MyoD(+) progenitors during the course of myogenic progression. Further investigation will be required to elucidate factors regulating MyoD-expressing cells.
In summary, we demonstrated for the first time that there are Myf5- and MyoD-expressing myogenic subpopulations during embryonic tongue myogenesis. The canonical Wnt signaling pathway differentially regulates these 2 subpopulations during tongue myogenesis. This study highlights the similarities and differences between tongue and limb muscle development and builds the foundation for a better understanding of the molecular regulatory mechanisms of tongue myogenesis.
Author Contributions
Z. Zhong, contributed to conception, design, and data analysis, drafted and critically revised the manuscript; H. Zhao, contributed to design and data analysis, drafted and critically revised the manuscript; J. Mayo, contributed to data analysis and interpretation, drafted and critically revised the manuscript; Y. Chai, contributed to conception, data acquisition, and analysis, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We thank Bridget Samuels for critical reading of the manuscript.
Footnotes
This study was supported by grants from the National Institute of Dental and Craniofacial Research, National Institutes of Health (R01 DE014078), to Yang Chai.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Biressi S, Molinaro M, Cossu G. 2007. Cellular heterogeneity during vertebrate skeletal muscle development. Dev Biol. 308:281–293. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 128:1253–1264. [DOI] [PubMed] [Google Scholar]
- Braun T, Gautel M. 2011. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 12:349–361. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. 2014. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 28:225–238. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. 2000. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 127:1671–1679. [DOI] [PubMed] [Google Scholar]
- Cusella-De Angelis MG, Molinari S, Le Donne A, Coletta M, Vivarelli E, Bouche M, Molinaro M, Ferrari S, Cossu G. 1994. Differential response of embryonic and fetal myoblasts to TGF beta: a possible regulatory mechanism of skeletal muscle histogenesis. Development. 120:925–933. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. 2008. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 135:1597–1604. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. 2007. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 11:375–388. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. 2008. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 14:437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Zhao H, Parada C, Hacia JG, Bringas P, Jr, Chai Y. 2012. A TGFbeta-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development. 139:1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R, Oka K, Yamaza T, Iwata J, Urata M, Xu X, Bringas P, Jr, Nonaka K, Chai Y. 2010. TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev Biol. 341:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. 2009. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 23:997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya M, Takada S. 1998. Wnt signaling from the dorsal neural tube is required for the formation of the medial dermomyotome. Development. 125:4969–4976. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. 1997. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 124:4729–4738. [DOI] [PubMed] [Google Scholar]
- Linker C, Lesbros C, Stark MR, Marcelle C. 2003. Intrinsic signals regulate the initial steps of myogenesis in vertebrates. Development. 130:4797–4807. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, et al. 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 22:1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo BS, Collins NS, Ashby P, Grigorieva E, Pevny LH, Candia A, Wright CV, Rigby PW, Pachnis V. 1999. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature 400(6739):69–73. [DOI] [PubMed] [Google Scholar]
- Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. 1991. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 111:1097–1107. [DOI] [PubMed] [Google Scholar]
- Otto A, Schmidt C, Patel K. 2006. Pax3 and Pax7 expression and regulation in the avian embryo. Anat Embryol (Berl). 211:293–310. [DOI] [PubMed] [Google Scholar]
- Parada C, Han D, Chai Y. 2012. Molecular and cellular regulatory mechanisms of tongue myogenesis. J Dent Res. 91:528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MH, Seale P, Rudnicki MA. 2003. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet. 4:497–507. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. 2005. Wnt signalling in stem cells and cancer. Nature. 434(7035):843–850. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 75:1351–1359. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Kuratani S, Tajbakhsh S. 2011. An eye on the head: the development and evolution of craniofacial muscles. Development. 138:2401–2415. [DOI] [PubMed] [Google Scholar]
- Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. 1989. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 341(6240):303–307. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Cordes R, Gossler A. 2007. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc Natl Acad Sci U S A. 104:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 143:134–144. [DOI] [PubMed] [Google Scholar]
- Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 21:70–71. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. 1998. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 125:4155–4162. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 127:5059–5070. [DOI] [PubMed] [Google Scholar]
- Voehringer D, Liang HE, Locksley RM. 2008. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol. 180:4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WM, Etemad S, Yamamoto M, Goldhamer DJ. 2013. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev Biol. 384:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. 2009. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 47:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.