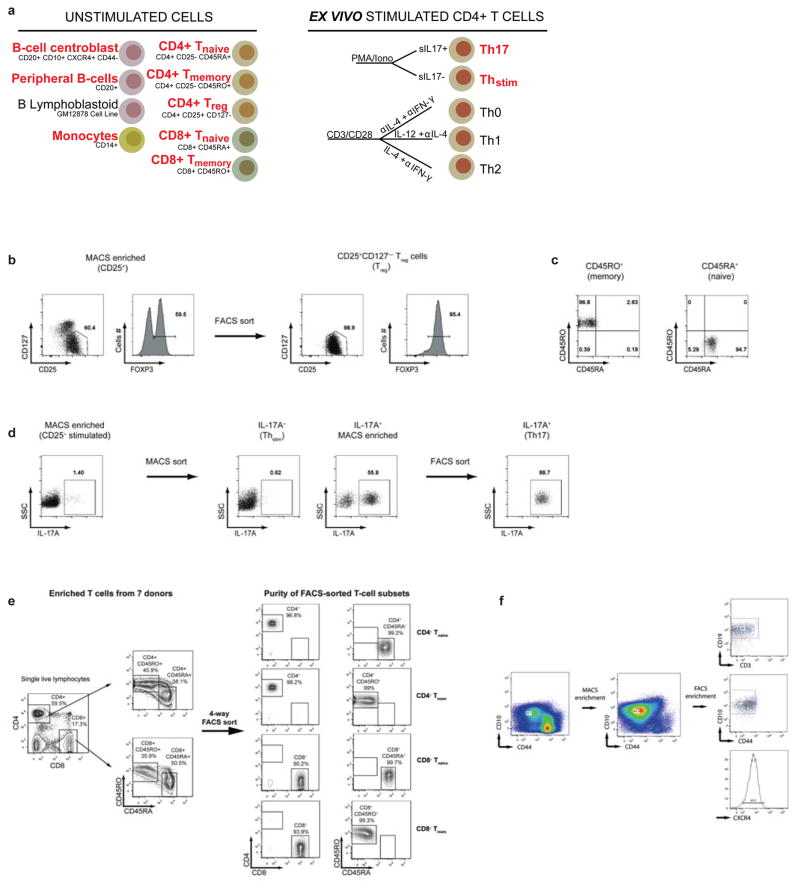
Extended Data Figure 6. Purification of Human Immune Cell Subsets.
(a) Immune populations subjected to epigenomic profiling in this study (red labels) or prior publications. (b) CD4+ cells were enriched based on CD25 expression (MACS) and subsequently sorted based on CD25highCD127low/− to isolate Treg cells; confirmed with FOXP3 intracellular staining. (c) CD4+CD25− cells were sorted to isolate Tmem (CD45RO+CD45RA−) and Tnaive (CD45RO−CD45RA+) cells. (d) CD4+CD25− cells were PMA/ionomycin stimulated and separated based on IL17 surface expression (MACS and FACS) to isolate Th17 cells (IL17+) and ThStim cells (IL17−). (e) Naïve (CD45RA+CD45RO−) and memory (CD45RA−CD45RO+) CD8+ T cells were isolated using a BD FACSAria 4-way cell sorter. Results are shown from one of two large-scale sorts. (f) Mononuclear cells were isolated from pediatric tonsils. Following CD10 enrichment (MACS), B centroblasts (CD19+CD10+CXCR4+CD44−CD3−) were purified by FACS.