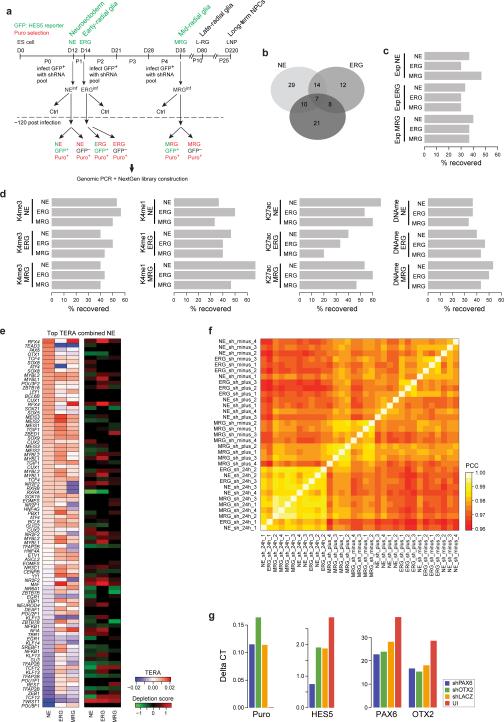
Extended Data Fig. 3 related to Fig. 3. Functional validation using a pooled shRNA screen.
a. Detailed outline of the pooled shRNA screen. Each stage (NE, ERG and MRG) was infected with an optimized virus titer aiming for an average of one shRNA integration per cell. Immediately after infection, cells were subjected to puromycin (puro) selection and bulk population material was collected 24h after infection and prior to efficient shRNA knockdown. Five days after infection and selection, cells were FACS sorted for HES5-GFP and both GFP+ and GFP- were collected for analysis. Subsequently, genomic DNA was extracted and all integrated shRNAs were amplified by PCR for each population separately. The resulting material was then used to construct libraries for next generation sequencing to count the number of shRNA integrations for each shRNA in each cell population.
b. Overlap of genes identified to facilitate HES5+ cell maintenance, progression or proliferation determined by genes with at least two shRNAs significantly (q≤0.05) overrepresented in the HES5+ population with respect to the 24h or HES5- control.
c. Regulator predictions based on differential gene expression. Performance is measured as percentage of the top 20 differentially expressed factors for each stage linked to the TF included in the shRNA library.
d. Regulator predictions based on TERA ranking for H3K4me3, H3K4me1, H3K27ac or DNAme. Performance is measured as percentage of the top 20 predicted activating or repressive motifs for each stage mapping to a TF included in the shRNA library.
e. Detailed heatmap showing the top 20 predicted motifs and corresponding TFs differentially active between the ES cell and NE stage based on the combined TERA scores for H3K27ac, H3K4me3, H3K4me1 and DNAme. In addition, knockdown results as depletion scores (green-red heatmap) obtained at each stage are shown on the right.
f. Heatmap showing the pairwise pearson-correlation coefficient (PCC) of the log2 read-count normalized shRNA libraries across all conditions and replicates.
g. Individual validation for shRNAs against OTX2 and PAX6 at the NE stage, which showed no effect in our pooled screening approach at any stage. Shown are qPCR levels for OTX2 or PAX6, HES5 and Puromycin relative to HPRT. Each gene was measured in an independent knockdown experiment for a pool of the 5 shRNAs against PAX6 (blue), OTX2 (green), lacZ (orange) as well as the uninfected control (red).