Abstract
Optogenetics has been recently applied to manipulate the neural circuits of Caenorhabditis elegans (C. elegans) to investigate its mechanosensation and locomotive behavior, which is a fundamental topic in model biology. In most neuron-related research, free C. elegans moves on an open area such as agar surface. However, this simple environment is different from the soil, in which C. elegans naturally dwells. To bridge up the gap, this paper presents integration of optogenetic illumination of C. elegans neural circuits and muscular force measurement in a structured microfluidic chip mimicking the C. elegans soil habitat. The microfluidic chip is essentially a ∼1 × 1 cm2 elastomeric polydimethylsiloxane micro-pillar array, configured in either form of lattice (LC) or honeycomb (HC) to mimic the environment in which the worm dwells. The integrated system has four key modules for illumination pattern generation, pattern projection, automatic tracking of the worm, and force measurement. Specifically, two optical pathways co-exist in an inverted microscope, including built-in bright-field illumination for worm tracking and pattern generation, and added-in optogenetic illumination for pattern projection onto the worm body segment. The behavior of a freely moving worm in the chip under optogenetic manipulation can be recorded for off-line force measurements. Using wild-type N2 C. elegans, we demonstrated optical illumination of C. elegans neurons by projecting light onto its head/tail segment at 14 Hz refresh frequency. We also measured the force and observed three representative locomotion patterns of forward movement, reversal, and omega turn for LC and HC configurations. Being capable of stimulating or inhibiting worm neurons and simultaneously measuring the thrust force, this enabling platform would offer new insights into the correlation between neurons and locomotive behaviors of the nematode under a complex environment.
INTRODUCTION
Comprehending the mechanosensory behavior of model animals, such as invertebrate nematode Caenorhabditis elegans (C. elegans), has been a critical task in model biology. In laboratory, C. elegans has been widely used as a model organism,1–3 due to its transparency, short life cycle, ease of maintenance, and genetic manipulation. In the context of brain and neuron related research, it is of great interest and significance to obtain a full map between environmental or external stimuli, sophisticated internal neural circuits and locomotive behavior. Compared to mammals like humans with neurons in the order of billions, C. elegans has only 302 neurons4 which are mostly well identified to facilitate further study. C. elegans' rather simple neuron circuits, on one hand, enable the nematode to exhibit behavior of higher-level animals. For example, the reversal activity of C. elegans in response to obstacles5 was studied and found to be related to multiple neurons with high complexity. On the other hand, they provide a controllable scale of neural network that could be manipulated.
Recent advances in microfluidics and microsystems6–14 have further facilitated investigation into C. elegans' behavior patterns in response to controlled external stimuli. Enabled by state-of-the-art technologies, a variety of external stimuli has been independently studied as a variable in the worm's environment, including temperature,15–17 chemicals,18–21 food-related signals,22 gas concentration,23 electric field,24,25 and physical obstacles.5,26 When input with these stimuli, the worm was generally treated as a black box to produce observable behaviors. By contrast, the role that internal neurons of C. elegans play was not yet explored in great depth, probably due to the lack of means to manipulate and image neurons in vivo. Nevertheless, many interesting attempts have been made in correlating external stimuli and locomotive behaviors. For example, a mathematical description of four typical C. elegans' locomotive patterns was proposed27 to formulate dynamics of the worm which freely crawls on an agar surface. In order to constantly characterize worm behaviors, micro-devices are deliberately designed to have functional modules which generate controllable stimulation on-chip such as electrodes for electric field,25 networked microchannels for olfactory signal delivery28 and chemical diffusion,18 polydimethylsiloxane (PDMS) gas-permeable membrane for oxygen gradient,23 and sinusoidal channels of different wavelengths for undulant locomotion.29
One limitation existing in most laboratory experiments, however, is that C. elegans is normally tested on a simple open and smooth planar substrate covered by agar or another medium, e.g., standard plate. This rather simple environment, though easy to implement, is insufficient to mimic the soil which is the natural habitat for C. elegans.30 In an attempt to provide a more soil-like environment, the concept of artificial soil30–32 was proposed, and “soil” of different forms was fabricated. Basically, the soil is composed of a large matrix of micro-pillars, in which the dimension and distance of pillars are varied. This idea was applied to study how C. elegans interacts with its surroundings. Interestingly, C. elegans was found to achieve enhanced locomotion with specific setting of the pillar structures,31 and particular neurons play a specific roles in crawling in structured environments.29 Furthermore, when made of materials, such as PDMS or SU-8, the pillars were capable of measuring subtle micro-Newton-scale mechanosensation forces between the worm body muscles and the pillars.32–34 Enabled by the force measurement techniques, the locomotion patterns could be precisely described quantitatively and correlated to environmental variables, which are favored by biologists. Meanwhile, it is still desirable to advance this further by incorporating means of manipulating neurons and thus provide a platform that allows mapping of external stimuli, internal neuron circuits, and locomotion patterns.
Very recently, optogenetics has emerged as an innovative and powerful technology for optical manipulation of neurons and animals.35–37 Together with precise position control of agile nematode, optogenetics has been applied to C. elegans to manipulate sensory neurons, interneurons, and motor neurons.20,38–44 Normally, certain neurons of interest in C. elegans can be selectively transferred with a gene that allows the neurons to be switched on/off when projected with light of specific wavelength (or color). In practice, as long as the color light is projected onto the worm body segment that possesses the neurons of interest, the neurons can be stimulated positively or negatively. This basic requirement thus facilitates the optogenetic manipulation of C. elegans neurons and leads to interesting findings. For instance, an original optogenetic system38 was applied to study the forward and backward activities coupled with two specific neurons, namely, AVA interneurons and PLM sensory neurons. The neural excitation level was quantified via the fluorescent image of neurons acquired by EMCCD (Electron-Multiplying CCD) and the result revealed the direct synchronization of AVA interneurons with the reversal from forward to backward motion, and the activity transients existing in PLM neurons are coupled to short spontaneous forward accelerations of the worm. In a different system,39 a liquid crystal display (LCD) projector was used along with laboratory microscopy and optics instead of expensive equipment to track a worm and project desired illumination patterns to worm body muscles for touch circuits. Nematodes were found to move in the form of triangles when the specific-frequency light is shed on the head and the tail, and in the form of S curve when the light is aimed on the belly. Similar to the previous systems, Kocabas et al.40 focused on AIY interneurons that are relevant to chemotactic behaviors. It was found that controlling of AIY was sufficient to force the animal to locate, turn towards and track virtual light gradients. Though these optogenetic systems are very powerful, they all used agar plate as the standard conditions for C. elegans living environment, which was mentioned earlier as not being able to sufficiently mimic the natural soil habitat.
In this paper, we report on the development of an optogenetic system integrating optical illumination for neuron manipulation and a microfluidic chip with soil-environment-mimicking and force-measuring micro-pillar arrays,33 taking advantage of both optogenetics and microfluidics. To integrate the optogenetic illumination, we used the same imaging and control architecture from earlier39 but different off-the-shelf components for the system due to the availability. We fabricated the micro-pillar PDMS array device as described previously.33 Since C. elegans is in a more complex pillar-structured array in this paper, we could not use the same image processing algorithm39,40 for C. elegans tracking and skeleton extraction. Therefore, we proposed a customized algorithm to isolate the worm from its surrounding pillars, and track and extract its skeleton automatically, based on which patterned illumination is generated and projected onto its different body segments. We have recently introduced the concept of a system combining optogenetics and environment-mimicking microsystem.45 In the following, we provide a full description of the integration and demonstrate its application to the collection of biologically relevant experimental data linking optical stimulation with force measurements and locomotive patterns. We believe that this integrated system holds the potential to function as an enhanced research platform, for the first time to our knowledge, to complete the information flow with capability of quantitative description between structured environment, internal neuron circuits, and locomotive behaviors.
METHODS
Microfluidic device design and fabrication
The device design and fabrication follows the exact procedures as described before.33 For completeness, the design and fabrication are briefly described here. As shown in Fig. 1(a), the device is composed of a matrix of PDMS micro-pillars extending from the bottom of a PDMS channel and a glass coverslip covering the top. Each pillar bends as a cantilever beam when the worm touches it in motion. To allow the free end of the beam to bend without unwanted interference, the sidewalls of the channel are designed 20 μm higher than the top of the pillars. This keeps the coverslip supported by the sidewall clear of the pillar tip. The channel has two open ends, one for loading of C. elegans and the other for nematode extraction. When the light source is switched on, structured illumination is projected onto specific body segments of the worm through the transparent glass lid.
FIG. 1.
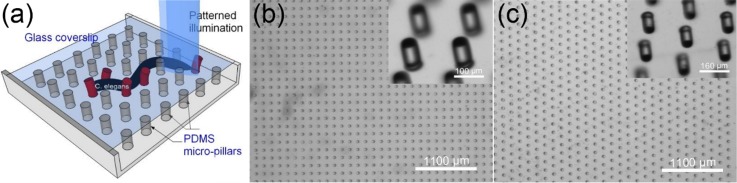
The micro-device. (a) Conceptual illustration of the device with PDMS micro-pillars mimicking the C. elegans environment and measuring mechanosensation force via those pillars in touch with the worm. Patterned illumination can be projected on selected segments of the worm body for neural manipulation. Top view of the PDMS pillars in lattice configuration (b) and honeycomb configuration (c) under optical microscope. Inset: zoom-in view of the micro-pillars.
To mimic the living environment, two different pillar configurations are designed as described before. These include a “lattice” (LC) design, in which the pillars are arranged in a square lattice grid structure (Fig. 1(b)), and a “honeycomb” (HC) design, in which the pillars are arranged in a hexagonal formation (Fig. 1(c)). The pillar dimensions are set to 100 μm height and 60 μm diameter for both configurations (see the inset in Figs. 1(b) and 1(c)). The micro-pillars are fabricated of PDMS using the standard soft lithography technique with procedures detailed previously.33 To allow sufficient space for the worm to move, the micro-pillar matrix spans 1 × 1 cm2. Figs. 1(b) and 1(c) show optical microscopy images of the pillar arrays for the two configurations. As the device is transparent for imaging and the micro-pillars each act as a vision-based force sensor, locomotion metrics such as the undulation frequency, amplitude, speed, and touch force can be retrieved via image processing in association with the optogenetic illumination.
Experimental setup
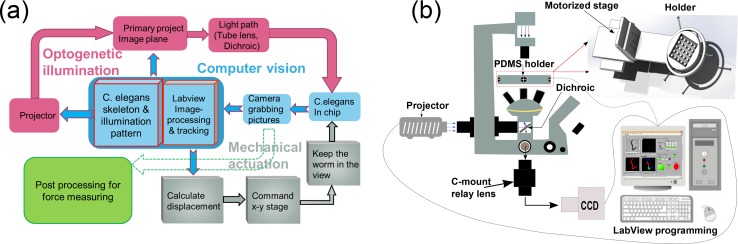
To incorporate the optogenetic illumination into the micro-device, the system ought to have four key modules in its architecture, as shown in Fig. 2(a). These modules essentially function to answer four basic questions. (i) What optogenetic illumination pattern to project? This question is answered by computer vision, whose core task is to constantly image the worm and extract its body segments, and subsequently generate a proper illumination pattern in association with the location of body segments that possess neurons of interest. (ii) Once one has the illumination pattern, how to project it onto C. elegans? This is addressed by a set of optical hardware devices, here collectively classified as optogenetic illumination, which convert the computer-generated light pattern from an artificial digital image to physical light, and project the light onto the nematode body via precise optics. (iii) Since the worm is actively moving, how to keep it in the field of view so that the previous questions are solvable? To answer this question, a module of mechanical actuation is required to physically bring the worm back in view in compensation for its instantaneous offset in motion. (iv) Finally, how to obtain its locomotion metrics? This question is then answered by the post-processing module previously reported,34 which analyzes time-stamped image sequences obtained in (i).
FIG. 2.
Experimental setup. (a) Overall system architecture of the platform. It includes four key modules: optogenetic illumination for neural manipulation, computer vision for illumination pattern alignment, mechanical actuation for C. elegans tracking, and post processing for offline force measurement. The blocks of the same module are labeled by identical color. (b) Schematic experimental setup of the platform. Note the microscope condenser provides bright-field light, the projector provides optogenetic light, the motorized stage actuates tracking of C. elegans, and the computer controls the hardware elements (e.g., camera, stage, and projector) and relevant image processing software.
Note that the first three modules work together as an online system, while the fourth module works offline. Therefore, it is necessary to synchronize the image sequences used in the offline processing and the illumination pattern image sequences generated online, which can be achieved by placing and comparing time stamps on the two image sequences. In the sequel, only the first three modules need embodiment in the experimental setup.
To integrate the optogenetic illumination, we use the same imaging and control architecture (Fig. 2(b))39 but different off-the-shelf components for the system due to availability. An inverted fluorescent microscope (Nikon Eclipse Ti-U) is used as a precise optical platform with readily available optical path for imaging and light projection. The standard fluorescent optical train is removed to free space for the installation of the custom optics, which allows an external projector to project optogenetic illumination onto the C. elegans sample. The light emitted from the projector horizontally passes through a tube lens (TI-BPU Back Port Unit) mounted on the back port of the microscope base, and is reflected upwards by a 705-nm dichroic (Semrock FF705-Di02) and focused by a 4× objective before reaching the sample. To image the sample, the microscope condenser provides bright-field illumination and the light passes the dichroic downwards, through built-in optics residing in the microscope base to be captured by a CCD camera (Basler Aca640-120 gm) connected to the side port. The worm sample is placed on the PDMS micro-device, which is firmly attached via a custom-made holder to a 3D Cartesian motorized stage (Sutter MP-285). Mounted on a custom support, the stage with 25 mm motion range in each direction can move at a resolution up to 40 nm and a speed up to 2.5 mm/s, satisfying the stringent requirement in both accuracy and speed to track the agile nematodes. Meanwhile, the projector, camera, and stage are all wired to a computer, which is equipped with customized LabVIEW algorithm and graphical user interface (GUI) to facilitate monitoring and control of the whole system. The setup, with exception of the computer, rests on an anti-vibration table (Zolix ZVB30-10) to minimize dynamic environmental noise.
Setting for optogenetic illumination
In our system, we choose a digital light processing (DLP) projector (Acer D101E, 2500 lumen) for pattern illumination due to availability and convenience. Both LCD and DLP projectors are able to generate sufficiently high-power light with acceptable contrast to activate the neurons, but DLP projectors are less expensive. Additionally, DLP projectors are not toned to be modified by adding extra optics in the light path to solve the convergence problem,39 which exists in LCD projectors when the projected image is too small such as is the case in our system. For certain color of illumination, its intensity is adjusted by varying the pixel values of RGB (Red, Green, and Blue) components. During the optical customization, two goals need to be met: (i) The object plane of the camera needs to overlap with the project image plane on the micro-device surface. This is to ensure that the two optical paths, one by the internal microscopy imaging system and the other by the external light projection system, can be identical in a range so when the worm is imaged in focus, a sharp illumination pattern can be projected onto its body. To do this, we adopt the “infinity-corrected” microscopy configuration,39 as shown in Fig. 3(a). Here, we remove the zoom lens of the projector, then place the projector in a position so that its primary project image plane is in a distance of focal length of the tube lens. On the other side, facilitated by the infinite space between the tube lens and the microscope objective, one has the freedom to place the dichroic mirror in the optical path, overcoming the restriction of limited physical space left by taking the fluorescence train out.
FIG. 3.
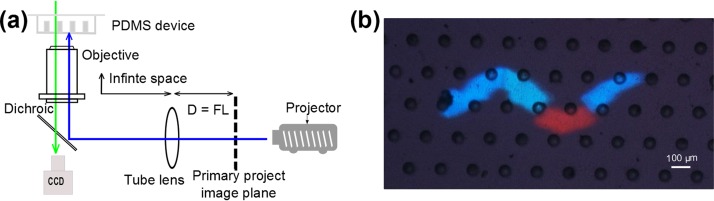
Optical setting. (a) Configuration for infinity-corrected microscope systems. (b) Snapshot of synchronized focus of the camera imaging and the projected pattern. Note the projected pattern resembles the contour of a worm body, but is not of identical dimension. Here, the picture was photographed by a color cooled digital camera DS-Ri1 to allow observation of the multi-color illumination patterns.
The second goal is to correctly align the camera image plane with the projector plane. This is to ensure the illumination pattern is projected onto the right segment of the worm body precisely by correcting the projection distortion caused by the inevitable misalignment of two optical paths in practice. Detailed procedures can be found elsewhere39 and are omitted here. Fig. 3(b) shows one example snapshot for an in-focus projected pattern and camera image of the pillar surface, showing good alignment.
Computer vision for generating illumination patterns
Image sequences of a moving worm during experiments are captured by the microscope camera. Based on the image sequences, the control system has two main tasks in image processing: (i) to visually identify and track a worm and (ii) to obtain the body skeleton of a worm so as to generate the color pattern in association with its different body segments. One common and key challenge for these two tasks is how to isolate the worm from its micro-pillar array background. To address this challenge, we propose a customized algorithm to isolate the worm body from its surrounding pillars. Before applying the algorithm, we down-sample the original-sized image (Fig. 4(a)) by reducing both its length and width by half. By doing so, small noise is removed from the original image and the computational burden for the algorithm is reduced. Based on the down-sampled image, the first step is to threshold the gray-level image and then retains the greatest white object (Fig. 4(b)) to isolate the worm and touched pillars from the background. For this, our setup uses a local adaptive thresholding method.46 To isolate the worm from its touching pillars (starting from Fig. 4(b)), we select the small, black, and isolated objects representing the pillars and then take inverse transformation to obtain the resultant image, as shown in Fig. 4(c). Furthermore, black-holes shown in Fig. 4(b) are filled, leading to the image shown in Fig. 4(d). After subtracting Fig. 4(c) from Fig. 4(d), a well-defined white object representing the isolated worm body is obtained.
FIG. 4.

Main steps for the customized image processing algorithm. (a) The original image. (b) The greatest white object selected from the binary image obtained by a local thresholding method. (c) Pillars separated from the greatest white object. (d) Black-holes in the greatest white object are filled. (e) Isolated worm body after subtracting (c) from (d). (f) Extracted and smoothed skeleton of the worm body, which is then divided into (e.g., four) segments. (g) Generated optogenetic illumination pattern using information in (e) and (f). Note the color for each segment can be varied or switched on/off at will.
Using this object, the worm is tracked through its centroid, and the skeleton extract from one end (head) to the other (tail). Compared to other work,39,40 the worm body is not perfectly smooth due to the contact between the body and pillars, resulting in a skeleton with some false branches. Therefore, the skeleton is further smoothed47 so as to keep only the longest (main) one among multiple branches. This is processed through MATLAB script and takes on average 0.02 s on our PC (3.10 GHz CPU, 8 GB memory), having insignificant effect on the computation speed. Subsequently, along the clean skeleton of the worm, its body is divided into certain number (i.e., four in this paper) of segments (Fig. 4(f)). For each segment, the desired color can be generated and the whole illumination pattern (Fig. 4(g)) is thus formed and sent to the projector for display.
In this paper, as the background is more complex, the entire image processing in the LabVIEW-based GUI control software takes about 70 ms for one loop, during which the worm may have moved 10.5 μm at a typical crawling speed of 150 μm/s (Ref. 33) before the stage catches up with the worm. To compensate for this offset, we deliberately set the illumination pattern greater than the contour of the worm.
Experimental methods
Before experiments, the PDMS surface was rendered hydrophilic through oxygen plasma treatment by a corona wand (Electro-Technic Products), and then moisturized by several drops of de-ionized (DI) water to provide an amenable living environment for C. elegans. The device was then mounted on the holder which is attached to the motorized stage, and the initial position of the stage was carefully adjusted so that later the stage can be motorized to cover the whole pillar-area. Once ready, a single worm was transferred carefully via a platinum probe from the nematode growth medium (NGM) to the micro-pillar device through the loading area, and then given at least 5 min to acclimatize to the pillar structure. Next, the stage is manually moved so that the worm is brought into the field of view. The software is commanded via the GUI to move the stage automatically by visual-tracking of the worm, and the illumination was on-the-fly switched on/off with proper settings of intensity and pattern. The illumination “on” time can be technically adjusted with no upper bound, but normally under 1 min (i.e., at seconds to 10-s scale) according to literature.39 In the course of automatic tracking and light projecting, the worm was also filmed and the video clips were analyzed offline for contact force and locomotion pattern analysis.
Our long-term goal is to collect ample of data for conclusive biological findings. In this paper, however, we aim to demonstrate the successful running of the integrated system and its application in biologically relevant data collection. Therefore, simply using wild-type C. elegans in experiment, we showcased adult worm for demonstration of illumination and force measurement in LC design, and L4 stage worm for observation of locomotion patterns under illumination in LC and HC designs. Wild-type worms were Bristol strain (N2) in this work; cultivated under standard conditions and fed OP50 bacteria on NGM according to Brenner.1 All experiments were conducted at room temperature (20 °C).
RESULTS AND DISCUSSION
Force measurement for adult wild-type worm under white light illumination
Automatic tracking of nematode proceeded well to keep the worm inside the field of view by commanding the stage correctly. The illumination pattern was generated in line with the image processing results of the body segments and white light was commanded to be projected onto the head segment. Mostly, the body skeleton could be recognized clearly without significant artifacts and the white light pattern was correctly generated and projected onto the desired body segments. The saved image sequences were analyzed for force measurement offline33 and the results are presented here.
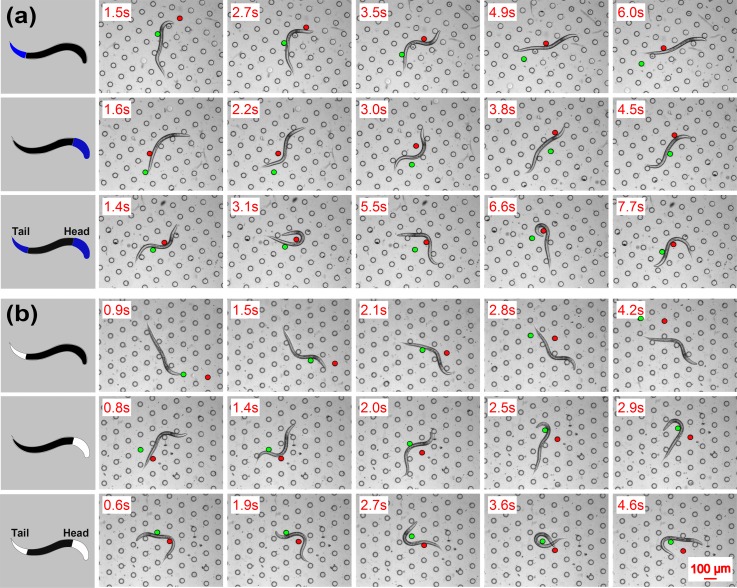
Fig. 5 shows several time-elapsed image sequences of a moving C. elegans under white light illumination onto its head segment. In each sequence, to better indicate the illustration pattern and location, we rendered false-color representing the white light pattern generated by the algorithm onto the head segment. To demonstrate the force measurement, we plot four circles representing four example pillars that engaged with different worm body segments during the imaging time, and superimpose the forces with magnitude and direction on top of the contact point. A 3.5 s long video clip was processed in the same manner.48
FIG. 5.
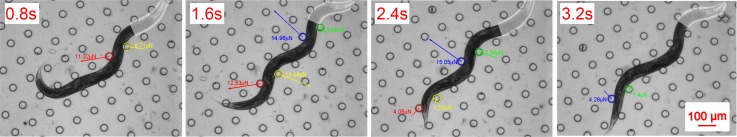
A wild-type C. elegans moves in the micro-pillar array under white light projected on its anterior (head) body segment. Pillar center-to-center distance is 140 μm. The white false-color represents white light projected on the worm head area. Different color-outlined circles are in contact with different worm body segments. The arrows pointing from the circle centers plot the force magnitude and direction.
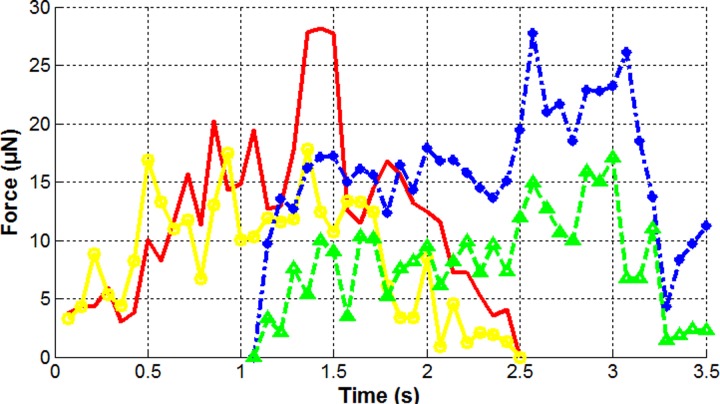
The time-elapsed force magnitude for the four pillars during the 3.5 s period is shown in Fig. 6. Compared to the case reported earlier,33 where there was no extra white light projected on the head segment, the average force was ∼2× greater and the maximum force level was ∼1.3× greater in this experiment for the same LC design. We hypothesize that this increase in force level is caused by the observed enhanced energetic activity of the worm under intensified illumination.49,50 In other words, when a worm attempts to avoid strong radiation such as 2500 lumen white light, it tries to excite its body muscles to escape the spot, therefore exerting greater force levels. Meanwhile, the maximum force always occurred when the middle part of the worm body contacted with the sensing pillar, such as reported for the case when no light was projected,33,51 suggesting that the worm inherently has the strongest muscles in that part and uses them regardless of the environmental situation.
FIG. 6.
Time-elapsed force measurement of the four selected pillars (identified by color) for a period of 3.5 s during which white light was projected on the worm's head.
Each curve in Fig. 6 represents the time-elapsed instantaneous touching force generated by the moving body of the sample worm. It shows rather significant variations compared to the reported data earlier,13 which measured the force at 5 Hz. According to our force measurement method,13,34 the significant variations between the data points in each curve are not mainly attributed to measurement noises. We believe these are the real signal variations. Actually, in the work of Doll et al.,32 higher-degree variations were observed in their experiment where the force sensing frequency was much higher than 14 Hz, which was used in our experiment. This indicates that a higher sensing frequency is able to reveal more detailed or fluctuating force information, a common sense in the field of signal processing.
Locomotive behavior of L4 stage wild-type worm under blue and white light illumination
To demonstrate that our system is applicable to smaller subjects, we also used early L4 stage worms which are less than half the size of the adult worms. Due to the smaller size, the force level was too low to be sensed by the pillars, but the worm tracking and light illumination worked well. Because the uncultivated worm exhibits a high sensitivity of avoidance to shortwave beaming, especially blue or ultraviolet light according to previous analysis,49,50 we first projected blue light to the head/tail segments and observed its locomotion patterns in the LC design. Then, we projected white light and observed its locomotion patterns in HC design, with the main aim to demonstrate the system's capability of providing different environments and illumination patterns.
Fig. 7(a) shows three representative locomotion patterns, including forward crawling, reversal, and omega turn under different blue light illumination patterns, in an attempt to avoid light radiation. As a general trend, with blue light illuminating its tail segment, the worm mainly exhibited forward crawling pattern; with blue light on its head segment, the worm exhibited reversal pattern mainly; when blue light on both head and tail segments, the worm exhibits omega turn mainly, but also forward crawling and reversal. A video clip showing these three patterns is available.48 When only standard microscopy illumination is used in structured environments,29,52 these three locomotion patterns also co-exist but occur in a more stochastic manner, different from the noticeable trends when the extra blue light applies. We speculate the specific illumination patterns play a role in exciting certain neurons in the head and tail segments of the worm. For example, AIY interneurons residing in the head segment were found related to reversals during foraging behavior through optical controlling of neurons.40,53 Furthermore, using laser ablation of neurons, two anterior neurons AVA and AVB interneurons residing in the head segment were found related to reversals; and posterior neuron PLM mechanosensory neurons residing in tail segment were found related to forward crawling.54 It appears that neurons in the head segment are responsible for reversals and the neurons in the tail for forward crawling, while neurons from both segments coordinate together to generate omega turns.
FIG. 7.
Three representative locomotion patterns observed under different conditions. Pillar center-to-center distance is 140 μm. (a) Blue light and LC device. (b) White light and HC device. The first row: Forward movement when blue light on tail segment. The second row: Reversal when blue light on tail segment. The third row: Omega turn when blue light on head and tail segments. Note that in each row, we randomly selected the same set of two pillars as reference to indicate the worm's locomotion.
We then changed the blue light to white, and changed the LC device to HC device, but projected light onto the same head/tail segment. Note here, we altered two factors simultaneously since we mainly aimed to demonstrate the system's capability. If, however, the main aim is to compare the two experiments with biologically conclusive data, one needs to follow strictly the control variate method. Under the current condition, Fig. 7(b) shows the similar locomotion patterns of the worm, for both white and blue light illuminations under LC and HC microstructures. This is not surprising as the blue light is simply one component of white light. For both light illuminations, we observed a time delay between the light projection and locomotion patterns in response to the projection. This observation is aligned with the previous report40 that neurons fire after a period of accumulation upon external stimulation. Though we currently are not able to quantify the delay, it will be of interest to do so in future once our system is equipped with an imaging apparatus for neuron activities.
Through the above-mentioned experiments, we have demonstrated the successful running of the integrated system for tracking and illuminating wild-type worms. The system is applicable for different stages of worms; able to generate and project desired illuminations of color, intensity, period, and pattern; and offers two configurations of environment-mimicking device. However, this work actually did not use any transgenetic worm in experiment to validate the effectiveness of optogenetics. There has been no direct evidence in genetics that neurons in N2 strain were excited. Therefore, it is more meaningful to use the transgenetic worm and verify the feasibility of the established method. This will be our future work.
CONCLUSIONS
We have fabricated PDMS micro-pillar array devices with two different configurations to mimic C. elegans habitat environment. To enable optogenetic illumination of C. elegans neurons and measure the corresponding thrust force, we incorporated two optical pathways in an existing inverted microscope. Together with the hardware setup, the system is equipped with four key modules accomplishing optogenetic illumination generation, illumination projection, automatic tracking of C. elegans, and force measurement. Specific optical settings have been configured, and customized image processing algorithms have been proposed to isolate the worm from the surrounding pillars and extract the skeleton. A LabVIEW-based GUI software has been programmed to support data collection. Using wild-type worms, we demonstrated the capability of the system in optical manipulation of C. elegans neurons, corresponding force measurement and observation of representative locomotive patterns. We envision that the integrated platform can serve as a powerful enabling tool for interrogation of neuron activities when C. elegans moves in its natural habitat-like environment.
ACKNOWLEDGMENTS
This work was supported by the NSFC (No. 61376120), the One-Thousand Young Talent Program of China, the National Instrumentation Project (No. 2013YQ19046701), and the MacDiarmid Institute for Advanced Materials and Nanotechnology.
References
- 1.Brenner S., “ The genetics of Caenorhabditis elegans,” Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulston J. E., Schierenberg E., White J. G., and Thomson J. N., “ The embryonic cell lineage of the nematode Caenorhabditis elegans,” Dev. Biol. 100, 64–119 (1983). 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- 3.Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., and Mello C. C., “ Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans,” Nature 391, 806–811 (1998). 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 4.Sternberg P. W., “ Working in the post-genomic C. elegans world,” Cell 105, 173–176 (2001). 10.1016/S0092-8674(01)00308-7 [DOI] [PubMed] [Google Scholar]
- 5.Nam S. W., Qian C., Kim S. H., van Noort D., Chiam K. H., and Park S., “ C. elegans sensing of and entrainment along obstacles require different neurons at different body locations,” Sci. Rep. 3, 3247 (2013) 10.1038/srep03247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crane M. M., Chung K., Stirman J., and Lu H., “ Microfluidics-enabled phenotyping, imaging, and screening of multicellular organisms,” Lab Chip 10, 1509–1517 (2010). 10.1039/b927258e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanik M. F., Rohde C. B., and Pardo-Martin C., “ Technologies for micromanipulating, imaging, and phenotyping small invertebrates and vertebrates,” Annu. Rev. Biomed. Eng. 13, 185–217 (2011). 10.1146/annurev-bioeng-071910-124703 [DOI] [PubMed] [Google Scholar]
- 8.San-Miguel A. and Lu H., “ Microfluidics as a tool for C. elegans research,” WormBook (September 24, 2013), edited by elegans The C.Research Community, WormBook; 10.1895/wormbook.1.162.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai X., Zhuo W., Liang Q., McGrath P. T., and Lu H., “ A high-throughput device for size based separation of C. elegans developmental stages,” Lab Chip 14, 1746–1752 (2014). 10.1039/c3lc51334c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J. and Wheeler A. R., “ Maze exploration and learning in C. elegans,” Lab Chip 7, 186–192 (2007). 10.1039/b613414a [DOI] [PubMed] [Google Scholar]
- 11.Aubry G. and Lu H., “ A perspective on optical developments in microfluidic platforms for Caenorhabditis elegans research,” Biomicrofluidics 8, 011301 (2014). 10.1063/1.4865167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo W. J., Sie Y. S., and Chuang H. S., “ Characterizations of kinetic power and propulsion of the nematode caenorhabditis elegans based on a micro-particle image velocimetry system,” Biomicrofluidics 8, 024116 (2014). 10.1063/1.4872061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benhal P., Chase J. G., Gaynor P., Oback B., and Wang W., “ Ac electric field induced dipole–based on-chip 3d cell rotation,” Lab Chip 14, 2717–2727 (2014). 10.1039/c4lc00312h [DOI] [PubMed] [Google Scholar]
- 14.Jin D., Deng B., Li J. X., Cai W., Tu L., Chen J., Wu Q., and Wang W. H., “ A microfluidic device enabling high-efficiency single cell trapping,” Biomicrofluidics 9, 014101 (2015). 10.1063/1.4905428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krajniak J. and Lu H., “ Long-term high-resolution imaging and culture of C. Elegans in chip-gel hybrid microfluidic device for developmental studies,” Lab Chip 10, 1862–1868 (2010). 10.1039/c001986k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang H. S., Chen H. Y., Chen C. S., and Chiu W. T., “ Immobilization of the nematode caenorhabditis elegans with addressable light-induced heat knockdown (ALINK),” Lab Chip 13, 2980–2989 (2013). 10.1039/c3lc50454a [DOI] [PubMed] [Google Scholar]
- 17.Hwang H., Krajniak J., Matsunaga Y., Benian G. M., and Lu H., “ On-demand optical immobilization of Caenorhabditis elegans for high-resolution imaging and microinjection,” Lab Chip 14, 3498–3501 (2014). 10.1039/C4LC00697F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung K., Zhan M., Srinivasan J., Sternberg P. W., Gong E., Schroeder F. C., and Lu H., “ Microfluidic chamber arrays for whole-organism behavior-based chemical screening,” Lab Chip 11, 3689–3697 (2011). 10.1039/c1lc20400a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht D. R. and Bargmann C. I., “ High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments,” Nat. Methods 8, 599–605 (2011). 10.1038/nmeth.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsch J., Ventimiglia D., Bargmann C. I., and Albrecht D. R., “ High-throughput imaging of neuronal activity in Caenorhabditis elegans,” Proc. Natl. Acad. Sci. U.S.A. 110, E4266–E4273 (2013). 10.1073/pnas.1318325110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi W., Wen H., Lu Y., Shi Y., Lin B., and Qin J., “ Droplet microfluidics for characterizing the neurotoxin-induced responses in individual Caenorhabditis elegans,” Lab Chip 10, 2855–2863 (2010). 10.1039/c0lc00256a [DOI] [PubMed] [Google Scholar]
- 22.Kopito R. B. and Levine E., “ Durable spatiotemporal surveillance of Caenorhabditis elegans response to environmental cues,” Lab Chip 14, 764–770 (2014). 10.1039/c3lc51061a [DOI] [PubMed] [Google Scholar]
- 23.Gray J. M., Karow D. S., Lu H., Chang A. J., Chang J. S., Ellis R. E., Marletta M. A., and Bargmann C. I., “ Oxygen sensation and social feeding mediated by a C. Elegans guanylate cyclase homologue,” Nature 430, 317–322 (2004). 10.1038/nature02714 [DOI] [PubMed] [Google Scholar]
- 24.Rezai P., Salam S., Selvaganapathy P. R., and Gupta B. P., “ Effect of pulse direct current signals on electrotactic movement of nematodes Caenorhabditis elegans and Caenorhabditis briggsae,” Biomicrofluidics 5, 044116 (2011). 10.1063/1.3665224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carr J. A., Parashar A., Gibson R., Robertson A. P., Martin R. J., and Pandey S., “ A microfluidic platform for high-sensitivity, real-time drug screening on C. Elegans and parasitic nematodes,” Lab Chip 11, 2385–2396 (2011). 10.1039/c1lc20170k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parashar A., Lycke R., Carr J. A., and Pandey S., “ Amplitude-modulated sinusoidal microchannels for observing adaptability in C. Elegans locomotion,” Biomicrofluidics 5, 24112 (2011). 10.1063/1.3604391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens G. J., Johnson-Kerner B., Bialek W., and Ryu W. S., “ Dimensionality and dynamics in the behavior of C. elegans,” PLoS Comput. Biol. 4, e1000028 (2008). 10.1371/journal.pcbi.1000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chronis N., Zimmer M., and Bargmann C. I., “ Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans,” Nat. Methods 4, 727–731 (2007). 10.1038/nmeth1075 [DOI] [PubMed] [Google Scholar]
- 29.Lockery S. R., Lawton K. J., Doll J. C., Faumont S., Coulthard S. M., Thiele T. R., Chronis N., McCormick K. E., Goodman M. B., and Pruitt B. L., “ Artificial dirt: Microfluidic substrates for nematode neurobiology and behavior,” J. Neurophysiol. 99, 3136–3143 (2008). 10.1152/jn.91327.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barriere A. and Felix M. A., “ High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations,” Curr. Biol. 15, 1176–1184 (2005). 10.1016/j.cub.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 31.Park S., Hwang H., Nam S. W., Martinez F., Austin R. H., and Ryu W. S., “ Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment,” PLoS One 3, e2550 (2008). 10.1371/journal.pone.0002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doll J. C., Harjee N., Klejwa N., Kwon R., Coulthard S. M., Petzold B., Goodman M. B., and Pruitt B. L., “ Su-8 force sensing pillar arrays for biological measurements,” Lab Chip 9, 1449–1454 (2009). 10.1039/b818622g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johari S., Nock V., Alkaisi M. M., and Wang W., “ On-chip analysis of C. Elegans muscular forces and locomotion patterns in microstructured environments,” Lab Chip 13, 1699–1707 (2013). 10.1039/c3lc41403e [DOI] [PubMed] [Google Scholar]
- 34.Ghanbari A., Nock V., Johari S., Blaikie R., Chen X., and Wang W., “ A micropillar-based on-chip system for continuous force measurement of C. Elegans,” J. Micromech. Microeng. 22, 095009 (2012). 10.1088/0960-1317/22/9/095009 [DOI] [Google Scholar]
- 35.Kim T. I., McCall J. G., Jung Y. H., Huang X., Siuda E. R., Li Y., Song J., Song Y. M., Pao H. A., Kim R. H., Lu C., Lee S. D., Song I. S., Shin G., Al-Hasani R., Kim S., Tan M. P., Huang Y., Omenetto F. G., Rogers J. A., and Bruchas M. R., “ Injectable, cellular-scale optoelectronics with applications for wireless optogenetics,” Science 340, 211–216 (2013). 10.1126/science.1232437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoi S. K., Kim V. H., Huy N. M., Sow C. H., Ow Y. S., and Bettiol A. A., “ Passive optical separation and enrichment of cells by size difference,” Biomicrofluidics 4, 44111 (2010). 10.1063/1.3523057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esteve V., Berganzo J., Monge R., Martinez-Bisbal M. C., Villa R., Celda B., and Fernandez L., “ Development of a three-dimensional cell culture system based on microfluidics for nuclear magnetic resonance and optical monitoring,” Biomicrofluidics 8, 064105 (2014). 10.1063/1.4902002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben Arous J., Tanizawa Y., Rabinowitch I., Chatenay D., and Schafer W. R., “ Automated imaging of neuronal activity in freely behaving Caenorhabditis elegans,” J. Neurosci. Methods 187, 229–234 (2010). 10.1016/j.jneumeth.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 39.Stirman J. N., Crane M. M., Husson S. J., Wabnig S., Schultheis C., Gottschalk A., and Lu H., “ Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans,” Nat. Methods 8, 153–158 (2011). 10.1038/nmeth.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kocabas A., Shen C. H., Guo Z. V., and Ramanathan S., “ Controlling interneuron activity in Caenorhabditis elegans to evoke chemotactic behaviour,” Nature 490, 273–277 (2012). 10.1038/nature11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leifer A. M., Fang-Yen C., Gershow M., Alkema M. J., and Samuel A. D., “ Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans,” Nat. Methods 8, 147–152 (2011). 10.1038/nmeth.1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen Q., Po M. D., Hulme E., Chen S., Liu X., Kwok S. W., Gershow M., Leifer A. M., Butler V., Fang-Yen C., Kawano T., Schafer W. R., Whitesides G., Wyart M., Chklovskii D. B., Zhen M., and Samuel A. D., “ Proprioceptive coupling within motor neurons drives C. elegans forward locomotion,” Neuron 76, 750–761 (2012). 10.1016/j.neuron.2012.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Z. V., Hart A. C., and Ramanathan S., “ Optical interrogation of neural circuits in Caenorhabditis elegans,” Nat. Methods 6, 891–896 (2009). 10.1038/nmeth.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schrodel T., Prevedel R., Aumayr K., Zimmer M., and Vaziri A., “ Brain-wide 3d imaging of neuronal activity in Caenorhabditis elegans with sculpted light,” Nat. Methods 10, 1013–1020 (2013). 10.1038/nmeth.2637 [DOI] [PubMed] [Google Scholar]
- 45.Qiu Z., Tu L., Xue X., Zhu T., Volker N., Li Y., Liu X., and Wang W., “ Optogenetic manipulation of freely moving C. elegans in an elastomeric environment-mimicking and force-measuring chip,” in MicroTAS, San Antonio, Texas, USA, 2014. [Google Scholar]
- 46.Ghanbari A., Nock V., Wang W., Blaikie R., Chase J. G., Chen X., and Hann C. E., “ Force pattern characterization of C. elegans in motion,” in 15th International Conference on Mechatronics and Machine Vision in Practice ( IEEE, Auckland, New Zealand, 2008), pp. 634–639. [Google Scholar]
- 47.Wang W., Sun Y., Dixon S. J., Alexander M., and Roy P. J., “ An automated micropositioning system for investigating C. elegans locomotive behavior,” J. Assoc. Lab. Autom. 14, 269–276 (2009). 10.1016/j.jala.2008.12.006 [DOI] [Google Scholar]
- 48.See supplementary material at http://dx.doi.org/10.1063/1.4908595E-BIOMGB-9-024501 for video clips showing C. elegans force measurement and locomotion patterns in couple with light projection.
- 49.Edwards S. L., Charlie N. K., Milfort M. C., Brown B. S., Gravlin C. N., Knecht J. E., and Miller K. G., “ A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans,” PLoS Biol. 6, e198 (2008). 10.1371/journal.pbio.0060198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward A., Liu J., Feng Z., and Xu X. Z., “ Light-sensitive neurons and channels mediate phototaxis in C. Elegans,” Nat. Neurosci. 11, 916–922 (2008). 10.1038/nn.2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X. N., Sznitman J., Krajacic P., Lamitina T., and Arratia P. E., “ Undulatory locomotion of Caenorhabditis elegans on wet surfaces,” Biophys. J. 102, 2772–2781 (2012). 10.1016/j.bpj.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croll N. A., “ Behavioural analysis of nematode movement,” Adv. Parasitol. 13, 71–122 (1975). 10.1016/S0065-308X(08)60319-X [DOI] [PubMed] [Google Scholar]
- 53.Gray J. M., Hill J. J., and Bargmann C. I., “ A circuit for navigation in Caenorhabditis elegans,” Proc. Natl. Acad. Sci. U.S.A. 102, 3184–3191 (2005). 10.1073/pnas.0409009101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chalfie M., Sulston J. E., White J. G., Southgate E., Thomson J. N., and Brenner S., “ The neural circuit for touch sensitivity in Caenorhabditis elegans,” J. Neurosci. 5, 956–964 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4908595E-BIOMGB-9-024501 for video clips showing C. elegans force measurement and locomotion patterns in couple with light projection.