Figure 1.
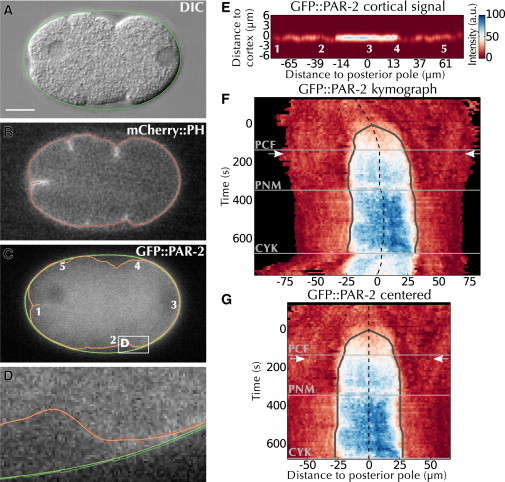
Quantifying polarity establishment. (A–D) Automated segmentation using ASSET. ASSET first (A) identifies the eggshell (green) in the DIC image, and then (B) segments the cell membrane (red) in the mCherry::PH image, and finally (C) transposes these two segmentations (green and red) to the GFP::PAR-2 channel and smoothes them (light green and orange). The white box delimits the area magnified in (D). Numbers in (C) and (E) denote corresponding locations. (D) Magnification of area delimited in (C) by the white box. (E) Perpendicular quantification of GFP::PAR-2 fluorescence in frame C (Fig. S1, H–M). (F) Kymograph of the recording containing the frame shown in (A)–(C) (see Movie S1), overlaid with automatic detection of the domain center (dashed line) and of domain expansion (gray line). Horizontal lines: cell cycle events (see Materials and Methods); arrows: position of the frame shown in (A)–(E). Black areas correspond to locations outside the embryo. (G) Final kymograph after centering and cropping, overlaid with landmarks as in (F). CYK, cytokinesis; PCF, pseudocleavage furrow; PNM, pronuclear meeting.To see this figure in color, go online.
