Figure 1. Datasets overview.
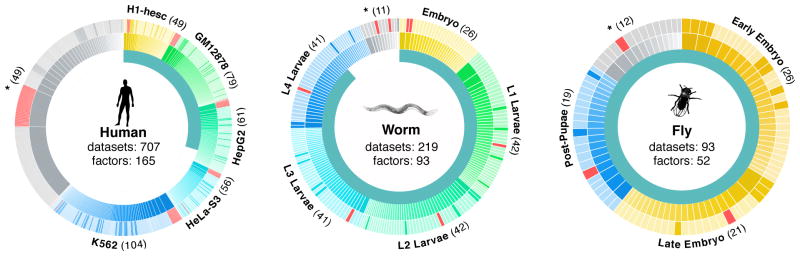
Data generated by the modENCODE and ENCODE consortium used in these analyses. The inner circle represents the fraction of datasets being presented for the first time in this paper. Each major context (cell lines in human and developmental stages in worm and fly) in each organism is colored a different hue in the outer two circles surrounding each organism and labeled on the edges of the diagrams. Datasets not in one of the main contexts are marked with asterisks. Each ChIP’d factor is depicted in the middle ring and the count is shown in parenthesis on the edges of the diagram (a given factor can be represented in multiple contexts). Every dataset is depicted in the outer ring, scaled by the number of peaks, and shaded to represent polymerase (red), transcription factor (lighter shade) and other (darker shade). In total 165, 93, and 52 unique factors were ChIP’d across all conditions and cell lines in human, worm, and fly respectively.
