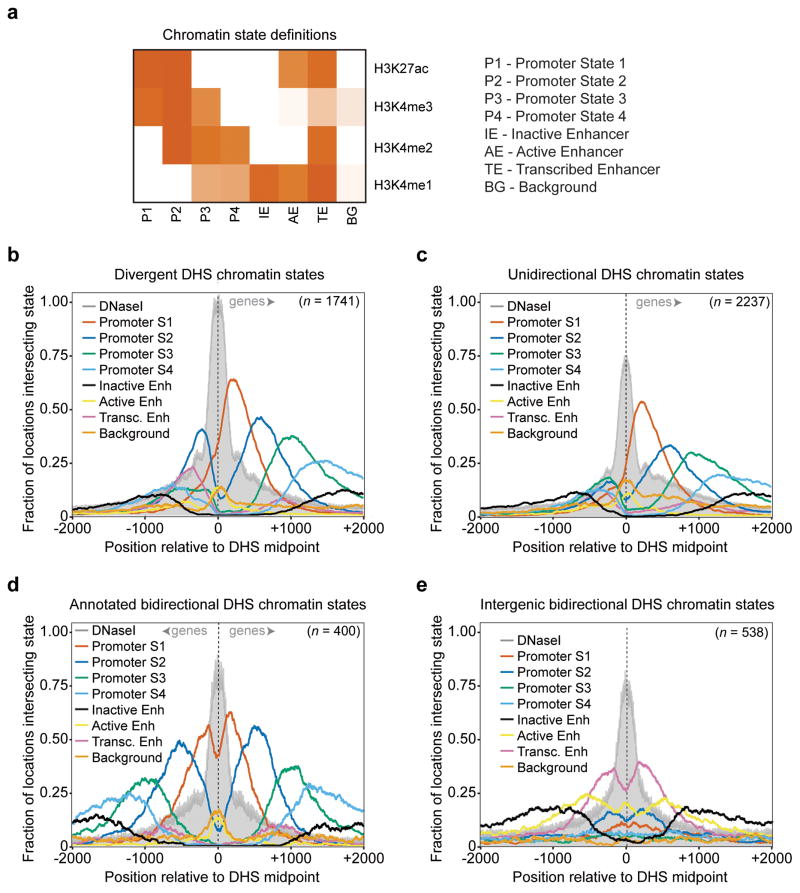
Figure 6. Distinct chromatin environment at unidirectional, bidirectional, and divergent promoter DHSs.
a, Chromatin state definitions based on Hidden Markov Model clustering of histone modification ChIP-Seq signal at 10 base pair resolution (see Experimental Procedures). Each state is a multivariate Gaussian distribution. Shown are the distribution mean vectors representing scaled, normalized ChIP-Seq signal. b, c, d, e, Chromatin state coverage ±2kb around the center of divergent promoter DHS (b), unidirectional promoter DHS (c), bidirectional promoter DHS (d), and divergent intergenic DHS (e) at single nucleotide resolution. Grey = DNaseI-seq read 5′end counts, red = Promoter State1, blue = Promoter State 2, green = Promoter State 3, light blue = Promoter State 4, black = Inactive Enhancer, yellow = Active Enhancer, pink = Transcribed Enhancer, orange = Background. See also Figure S6.