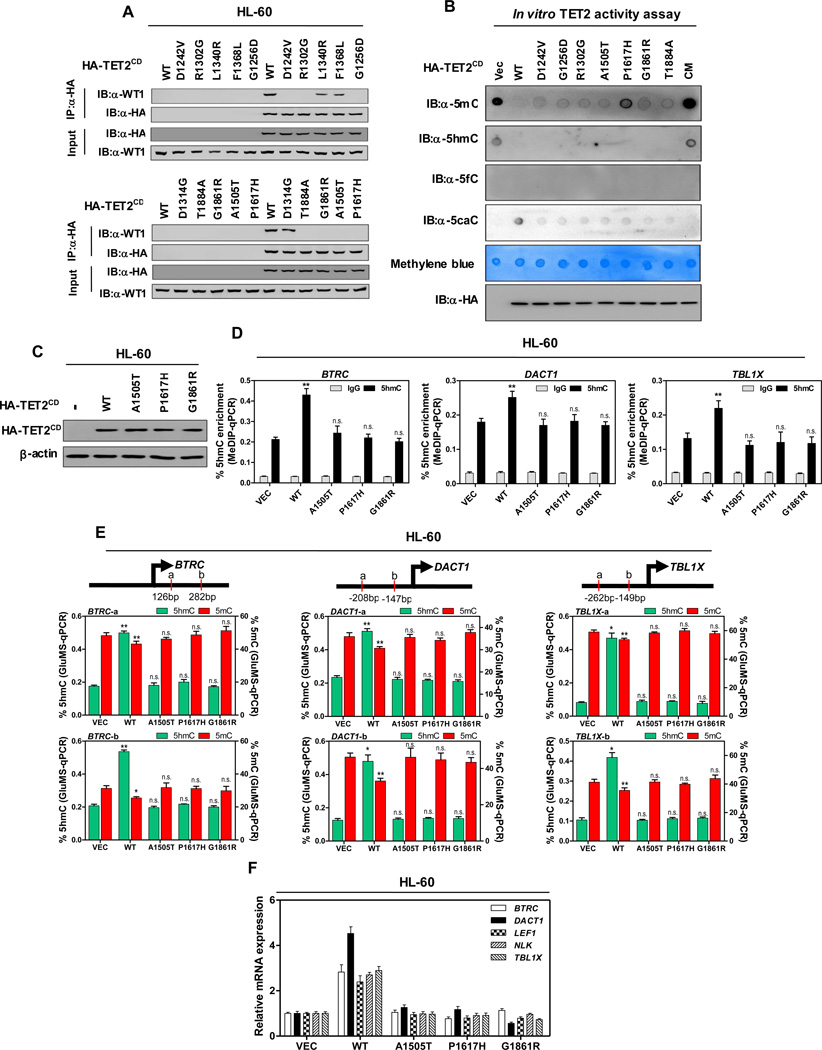
Figure 5. AML-derived mutations in TET2 disrupt WT1 binding.
(A) HL-60 cells were transiently transfected with vectors encoding HA-tagged wild-type TET2CD, or AML-derived WT1-binding defective TET2 mutants as indicated, and the ectopically expressed TET2 proteins were immunoprecipitated, following by western blot to detect endogenous WT1.
(B) In vitro TET2 catalytic activity assay. Genomic DNA were isolated from human monocyte-derived macrophage (MDM) cells, sonicated and incubated with immunopurified HA-tagged wild-type and mutants human TET2 at 37°C for 2 hrs. After termination of the reaction, a fraction of reaction mixture from each reaction was subjected to dot-blot assay using the antibodies specific for 5mC, 5hmC, 5fC and 5caC. See Experimental Procedures for more details. The amount of DNA in each reaction was examined by methylene blue staining. CM refers to a catalytic mutant of TET2 that harbors two mutations disrupting the binding with Fe2+
(C) HL-60 cells were transduced with lentiviral vectors encoding HA-tagged wild-type TET2CD, or AML-derived WT1-binding defective TET2CD mutants as indicated, and the expression of ectopic TET2CD proteins was determined by western blot.
(D–F) Stable HL-60 cells were generated as described above in (C). The 5hmC enrichment at TET2 binding sites at the promoter regions of indicated WT1-target genes was determined by hMeDIP-qPCR (D), rabbit IgG was included as negative control. Moreover, the site-specific levels of 5hmC and 5mC were determined by using GluMS-qPCR (E), and the mRNA expression of indicated WT1-target genes was determined by qRT-PCR (F).
Shown are average values of triplicated results with standard deviation (S.D.). *denotes the p < 0.05 and **denotes the p < 0.01 for the indicated comparison. n.s.= not significant.
See also Figure S5.