Figure 4.
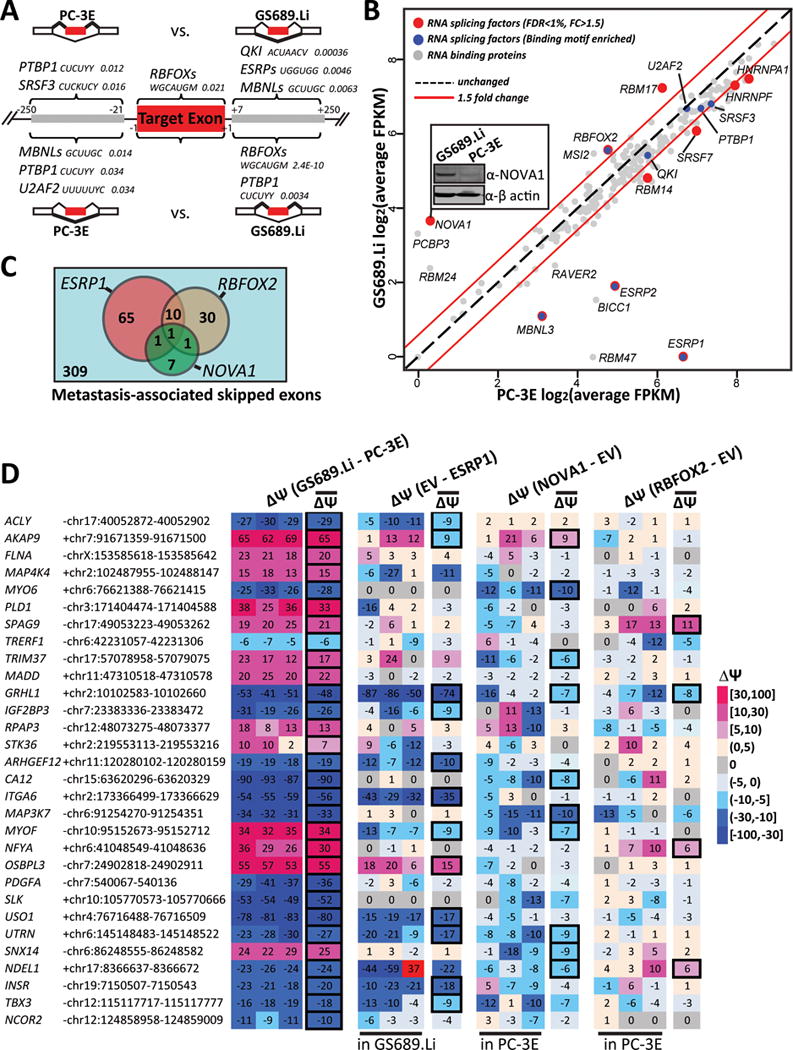
Splicing factors responsible for alternative splicing regulation associated with metastatic colonization.
(A) Significantly enriched binding sites of splicing factors and other RNA binding proteins in differential exon skipping events between PC-3E and GS689.Li. Gene symbols of splicing factors are followed by the consensus binding motifs and their Benjamini-adjusted p-values. Y: C/U; K: G/U; W: A/U; H: U/C/A; V: A/C/G; M: A/C. (B) Scatter plot of 221 RNA binding proteins’ average gene expression levels (log2 FPKM) in PC-3E and GS689.Li. Splicing factors with significant gene expression change are colored in red, while splicing factors whose binding motifs are significantly enriched are colored in blue. Some splicing factors belong to both categories. The inset image shows the western blot confirmation of NOVA1 protein expression levels. (C) The number of known target exons of ESRP1, RBFOX2, or NOVA1 among differential exon skipping events between PC-3E and GS689.Li. (D) Heatmap showing the effects on 30 randomly selected metastasis-associated skipped exons by ectopic expression of ESRP1, NOVA1, and RBFOX2. The Δψ values between PC-3E and GS689.Li or upon ectopic expression of a specific splicing factor are color coded for three individual replicates and the average of all three replicates. Significant splicing changes are highlighted by the bold box. EV: empty vector (control).
