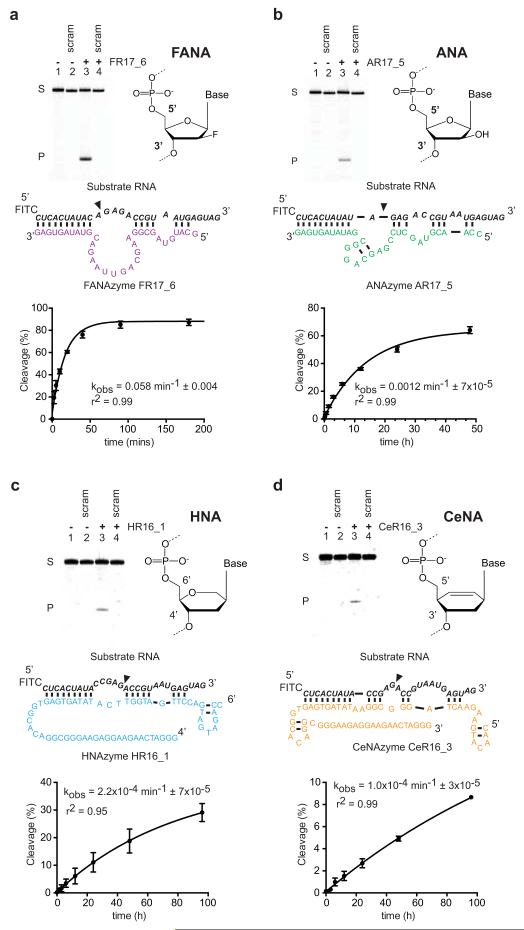
Figure 1. RNA endonuclease XNAzymes elaborated in four synthetic genetic polymer chemistries.
Gel electrophoresis, putative secondary structures and pre-steady state reaction rates (kobs) (n=3, error bars=sd, 25°C) of a, FANA, b, ANA, c, HNA, d, CeNA enzymes. Urea-PAGE gels show bimolecular cleavage in trans of cognate RNA substrates (NucSR variants, see Extended Data Fig. 3) (lanes 1 & 3), but not scrambled RNA (NucSR SCRAM1) (lanes 2 & 4), catalyzed by XNAzymes (lanes 3 & 4) (17°C).