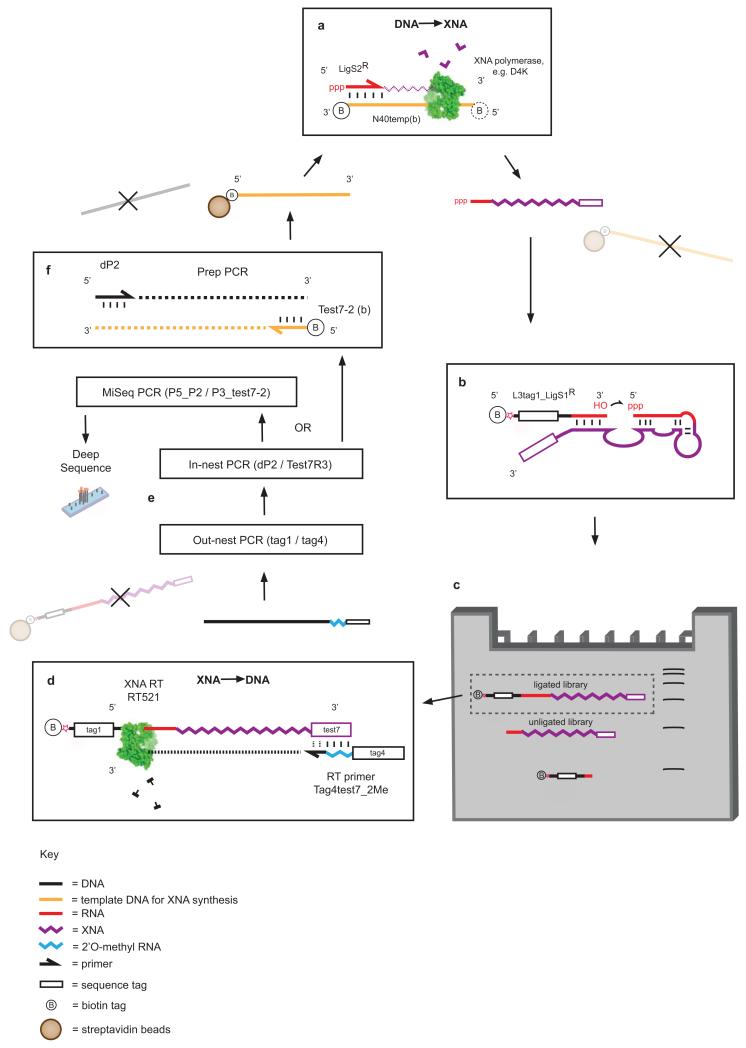
Extended Data Figure 6. Selection scheme for RNA ligase XNAzymes.
a, XNA library preparation using DNA-dependent XNA polymerases, primed by a 5′ triphosphorylated (5′ppp) RNA primer (LigS2R), which serves as one of the substrates for RNA ligation in cis. Libraries are synthesized with 3′ biotinylated DNA template, allowing capture and removal by streptavidin beads. b, Single-stranded libraries (unbiotinylated) are annealed and incubated in reaction buffer (see Method) together with a biotinylated chimeric DNA-RNA substrate (tag1_LigS1R), which successful XNAzymes ligate to RNA substrate LigS2R in cis. c, Size separation of reacted XNA pools using Urea-PAGE. Ligated XNA pools are gel-extracted and captured by streptavidin beads. d, Reverse transcription of XNA pools using XNA-dependent DNA polymerase RT521L, which is also able to transcribe RNA across the ligation junction (i.e. [RNA-RNA-XNA] -> cDNA). e, Amplification of transcribed cDNA by successive PCR reactions; out-nest reaction depends on priming site (tag1) from ligated substrate tag1_LigS1R. f, PCR reaction generating templates for XNA synthesis (now 5′ biotinylated) for further rounds of selection.