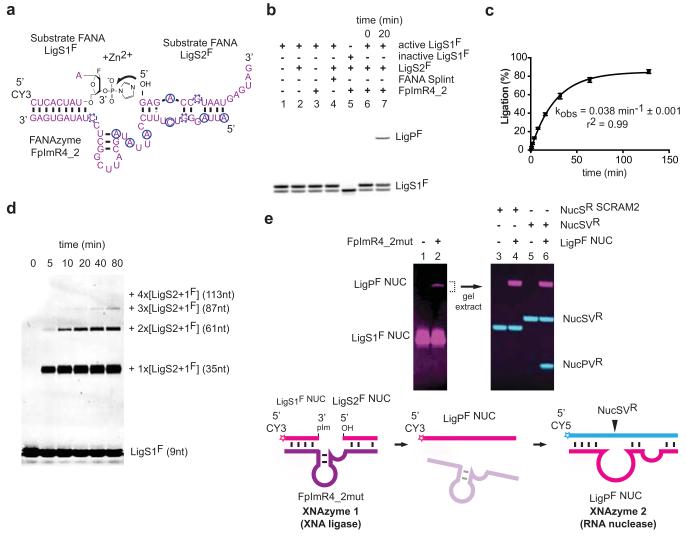
Figure 4. XNA-XNA ligase XNAzyme (FANA) demonstrates catalysis without natural nucleic acids.
a, Secondary structure (determined by DMS mapping, Extended Data Fig. 5) of chemically-synthesised FANAzyme FpImR4_2, which ligates FANA LigS1F, activated with 3′ phosphorylimidazolide (pIm), to LigS2F in trans. b, Urea-PAGE gel showing no product with: substrate LigS1F alone (lane 1), no XNAzyme (lane 2), no LigS2F (lane 3), splint (lane 4), or LigS1F lacking 3′pIm (lane 5); product formation is dependent on LigS2F, activated LigS1F and XNAzyme (lanes 6 and 7). c, Pre-steady state trimolecular reaction rate (kobs) (n=3, error bars=sd, 35°C). d, FpImR4_2-catalysed oligomerisation of XNA (FANA) substrates. e, XNAzyme-catalysed assembly of an active XNAzyme. A variant XNA ligase (FpImR4mut) catalyses ligation (lane 2) of FANA substrates LigS1F NUC and LigS2F NUC. The product (LigPF NUC) is a variant of XNAzyme FR17_6min (Fig. 2), which cleaves RNA substrate NucSVR (lanes 5 and 6), but not scrambled RNA (NucSR SCRAM2) (lanes 3 and 4).