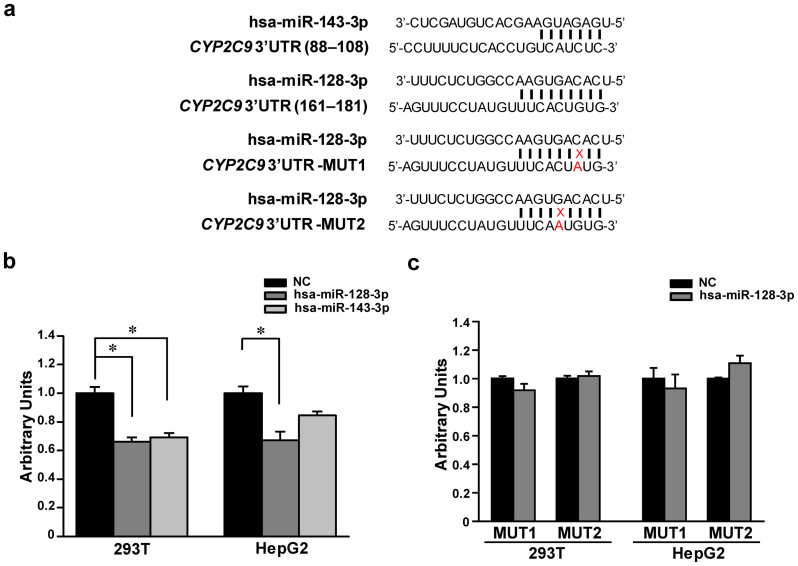
Figure 1. The hsa-miR-128-3p and hsa-miR-143-3p suppressed reporter gene expression.
(a) Prediction of hsa-miR-143-3p targeting to the 3′-UTR of CYP2C9, and hsa-miR-128-3p targeting to the 3′-UTR of CYP2C9, as well as the “targeting” of hsa-miR128-3p to two mutated 3′UTR sequences of the CYP2C9 gene. The solid vertical line indicates base pairing and the numbering used for the hsa-miR-128-3p and hsa-miR-143-3p target sequences (88–108 or 161–181, respectively) is consistent with that given for the CYP2C9 3′-UTR in NM_000771. (b) Luciferase reporter assays to investigate the effects of the hsa-miR-128-3p and hsa-miR-143-3p on CYP2C9 3′-UTR. (c) Luciferase reporter assays to investigate the effect of hsa-miR-128-3p on mutated CYP2C9 3′-UTRs. 293T and HepG2 cells were transiently transfected with the CYP2C9- CU, CYP2C9-MUT1-CU (containing a mutated sequence in the 3′UTR of CYP2C9), or CYP2C9-MUT2-CU (containing another mutated sequence in the 3′-UTR of CYP2C9) plasmid, together with 50 nmol/L hsa-miR-128-3p mimic, hsa-miR-143-3p mimic, or miRNA negative control, respectively, and harvested 48 hours after transfection. Renilla luciferase activity was measured, and then normalized to firefly luciferase. Three independent transfection experiments were carried out, and each was performed in triplicate. Data are shown as relative activity to luciferase activity expressed by the CYP2C9-CU plasmid transfection together with miRNA negative control. * P < 0.05; NC, miRNA negative control.