Figure 2.
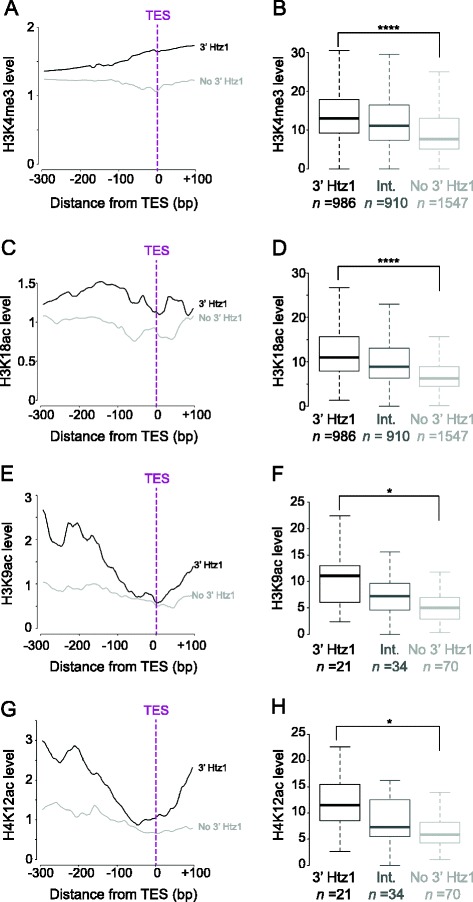
Htz1 at 3′ ends of genes co-localises with active histone modifications. Histone modifications known to occupy active promoters include H3K4me3 [19], H3K18ac [20], H3K9ac and H4K12ac [18]. H4K4me3 and H3K18ac data are from the whole genome; H3K9ac and H4K12ac are restricted to chromosome 3. A, C, E, G. Profiles of histone modifications at the 3' ends of genes with 3' Htz1 enrichment (black lines) or without Htz1 enrichment (grey lines). All four active histone marks are enriched upstream of the TESs of genes that have high 3' Htz1 occupancy. B, D, F, H. Boxplots of the distributions of histone modification levels in genes with high 3' Htz1 enrichment (3' Htz1; black boxes), without Htz1 enrichment (no 3' Htz1; light grey boxes) or with intermediate levels of 3' Htz1 (Int.; medium grey boxes). The number of genes in each category is indicated. H3K4me3 (p = 4.0 x 10−29), H3K18ac (p = 8.0 x 10−57), H3K9ac (p = 6.4 x 10−3) and H4K12ac (p = 1.2 x 10−2) are significantly higher on genes with high 3' Htz1. *p ≤ 0.05, ****p ≤ 0.0001. The p-values were obtained using two-tailed t-tests.
