Figure 1.
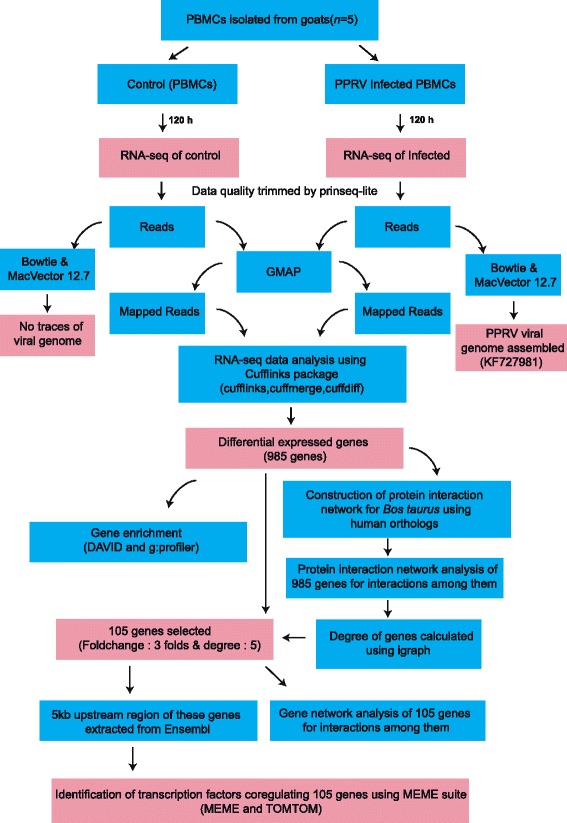
Overview of the workflow. PBMCs were collected from 5 goats and infected with PPRV virus at 1.0 mulitiplicity of infection (MOI). Uninfected PBMCs acted as control. RNA was isolated and sequenced from both infected and control PBMCs. The reads were quality filtered and mapped to the Bos taurus reference genome using GMAP program. Differential gene expression in infected vs control PBMCs was identified by subjecting the quality reads through the cufflinks package. Functional enrichment analysis was done using g:profiler and DAVID. Protein – protein interaction network was constructed for differentially expressed genes using the human reference interaction network and analyzed. Differentially expressed and highly connected (DHEC) 105 genes were selected and overrepresented conserved DNA motifs upstream of these genes were identified using MEME.TFs binding to these motifs were predicted using TOMTOM.
