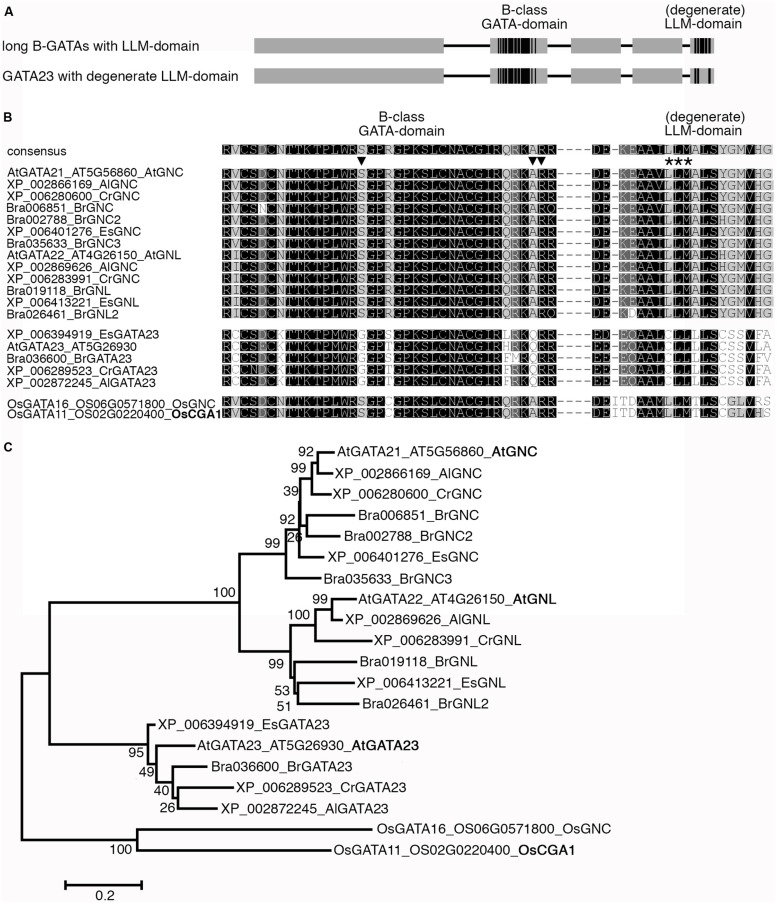
FIGURE 9.
GATA23 is specific for the Brassicaceae. (A) Schematic representation of long B-GATAs with the C-terminal LLM-domain and GATA23 with a degenerate LLM-domain. Boxes represent protein regions with sequence similarity (gray) or high sequence conservation (black), lines represent protein regions with restricted sequence conservation. (B) Sequence alignment of the GATA-domain and the (degenerate) LLM-domain of the LLM-domain containing B-GATAs AtGNC and AtGNL as representatives for LLM-domain containing B-GATAs as well as AtGATA23 from A. thaliana (At) as a B-GATA with a degenerated LLM-domain and their predicted orthologues from other Brassicaceae: A. lyrata (Al), Capsella rubella (Cr), Brassica rapa (Br), Eutrema salsugineum (Es). Whereas the core LLM-motif is conserved among the GNC and GNL orthologues from the different Brassicaceae species and rice, it is divergent in the GATA23 B-GATAs. The triangles mark characteristic amino acid residues of the B-GATA domain that allow predicting the presence of the LLM-domain or a degenerate LLM-domain. Please note the conservation of these residues between the LLM-domain containing B-GATAs from the Brassicaceae and rice whereas the GATA23 orthologues are also divergent in these residues in the DNA-binding domain. (C) Phylogenetic tree of the B-GATAs shown in (B). The phylogenetic tree was generated using the Geneious R7 Software based on a MUSCLE alignment in MEGA6.06 using the following settings: Gap penalty, gap open -2.9, gap extend 0, hydrophobicity multiplier 1.2; interations, maximum iterations 8; clustering method, all iterations UPGMB and minimum diagonal length (lambda) 24. The Neighbor Joining tree was generated with the bootstrap method (1000 replications) using the Jones-Taylor-Thornton model using default settings. Bootstrap values are indicated by each node. Bar = 0.2 amino acid substitutions per site.