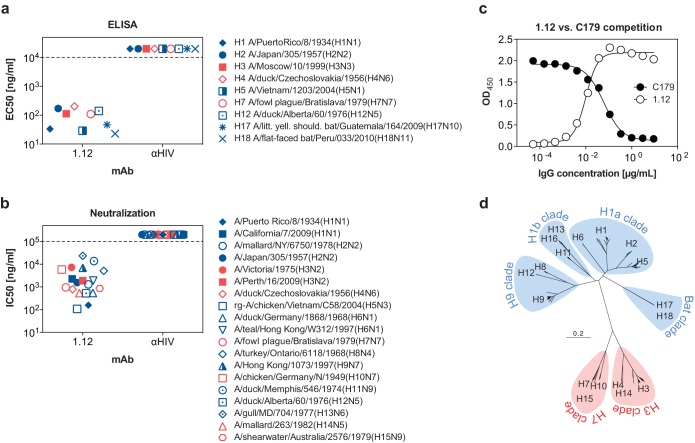
FIG 1.
Specificity of hMAb 1.12. (a) Half-maximal binding concentrations (EC50s) of hMAb 1.12 to recombinantly expressed HA proteins from 9 subtypes. The data represent the means of EC50s obtained in two independent experiments. (b) Half-maximal neutralizing concentrations (IC50s) of MAb 1.12 to a panel of 19 viruses from 15 subtypes. The HIV-1 gp120-specific MAb b12 was used as a negative control in both experiments (αHIV). A representative of at least 2 independent, consistent experiments performed in triplicate is shown. Avian viruses are depicted with open symbols, human isolates with solid symbols, and bat isolates with asterisk-like symbols. Isolates with zoonotic potential are designated with mixed symbols. Isolates from phylogenetic group 1 are depicted in blue and those of phylogenetic group 2 in red. A dashed line is used to indicate the detection limit of the assay. (c) The epitope recognized by hMAb 1.12 was roughly evaluated in a binding-competition ELISA using HA stem-reactive hMAb c179. ELISA plates coated with purified HA from A/Puerto Rico/8/1934(H1N1) were incubated with titrated amounts of hMAb 1.12, washed, and later incubated with a fixed concentration (1 μg/ml) of the murine hMAb c179. Binding of both antibodies was then detected using species-specific secondary antibodies. (d) Phylogenetic tree for all 18 HA subtypes. At total of 1,339 arbitrarily chosen recent nonidentical HA amino acid sequences (from the year 2000 to the present for frequent isolates; from 1985 to the present for rare isolates) were aligned using Muscle 3.8 (34). The tree was built using “neighbor” from the Phylip 3.69 software package (http://evolution.genetics.washington.edu/phylip.html) and illustrated as a rooted tree in FigTree 1.4 (http://tree.bio.ed.ac.uk/software/figtree). Phylogenetic group 1 is indicated in blue and group 2 in red.