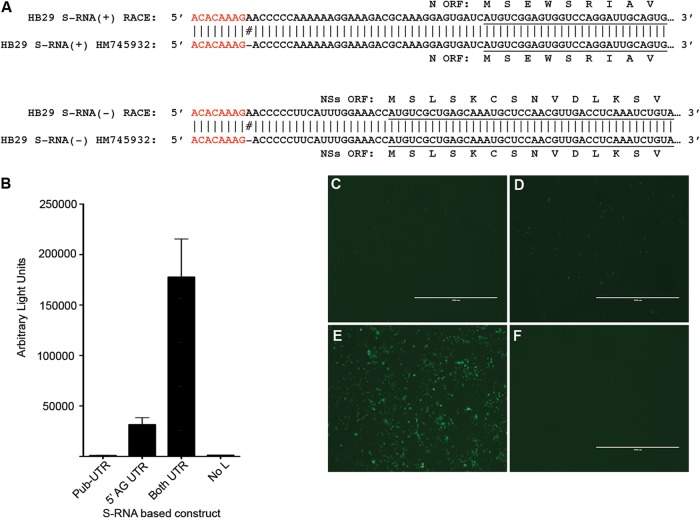
FIG 2.
Correction of the sequence of the S-segment-based minigenome. (A) Schematic representation of 3′ RACE data indicating that there is a missing A residue at position 9 of both the 5′ and 3′ S segment UTRs. Conserved terminal nucleotides are highlighted in red, and open reading frames for N and NSs proteins are underlined. (B) Effect of correcting the S-segment UTR sequence on S minigenome activity. BSR-T7/5 cells were transfected with pTM1-HB29N, pTM1-HB29ppL, pTM1-FF-Luc and contained either the published UTR sequences (Pub-UTR) or pTVT7-HB29SdelNSs:hRen with an A residue inserted at position 9 of the 5′ antigenomic UTR (5′AG UTR) or pTVT7-HB29SdelNSs:hRen with A and T residues inserted at position 9 of both 5′ and 3′ UTRs (Both UTR). “No L” is a negative control without pTM1-HB29ppL (empty pTM1 vector was used instead). Renilla luciferase activity was measured 24 h posttransfection. (C to F) eGFP autofluorescence observed in cells 24 h after transfection with analogous minigenome plasmids containing eGFP in place of hRen sequences: HB29 S with published UTR sequences (C); A residue inserted at position 9 of the 5′ antigenomic UTR (D); A and T residues inserted at position 9 of both 5′ and 3′ UTRs (E); negative control without pTM1-HB29ppL (F).