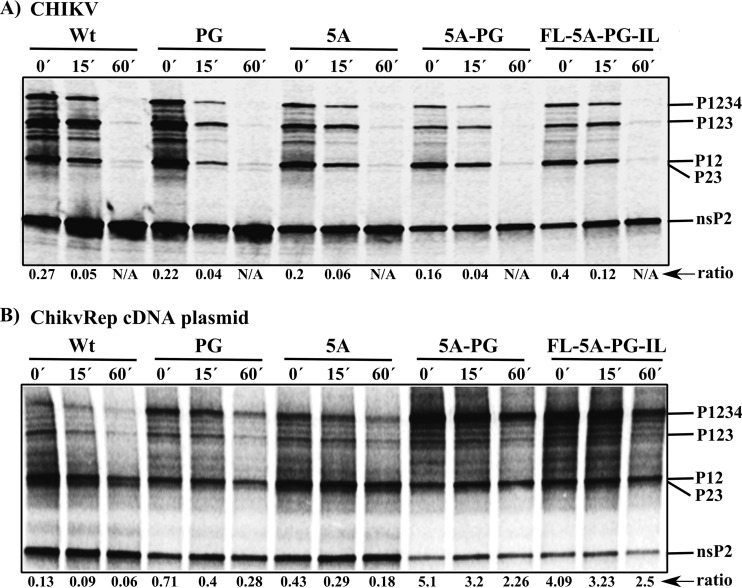
FIG 5.
Analysis of the proteolytic processing of wt and mutant nonstructural polyproteins. The names of viruses and replicon cDNA-containing plasmids used in the assays are indicated at the tops of the panels. The positions of the nonstructural polyproteins and nsP2 are indicated on the right side of the gels. (A) Processing of nonstructural polyproteins in virus-infected BHK-21 cells. BHK-21 cells were infected with wt CHIKV, CHIKV-PG, CHIKV-5A, CHIKV-5A-PG, or CHIKV-FL-5A-PG-IL at an MOI of 20 PFU/cell. At 3 h p.i., cells were starved in methionine-free and cysteine-free medium, labeled for 15 min with [35S]methionine-cysteine (pulse, 0′), and chased for 15 or 60 min in medium containing excess unlabeled methionine and cysteine. The cells were lysed in 1% SDS, and proteins were denatured by boiling and analyzed via immunoprecipitation with anti-nsP2 followed by SDS-PAGE and visualization with a Typhoon imager. (B) Processing of in vitro-translated wt and mutant nonstructural polyproteins. The reaction was carried out using the TNT SP6 rabbit reticulocyte system and plasmids containing the indicated ChikvRep cDNAs as templates. The produced nonstructural polyproteins and the products of their processing were denatured by boiling in 1% SDS. The samples were analyzed as described for panel A. The ratio is the amount of label that was incorporated into unprocessed P1234 compared with the amount incorporated into individual nsP2 protein. Results from one of two reproducible experiments are shown.