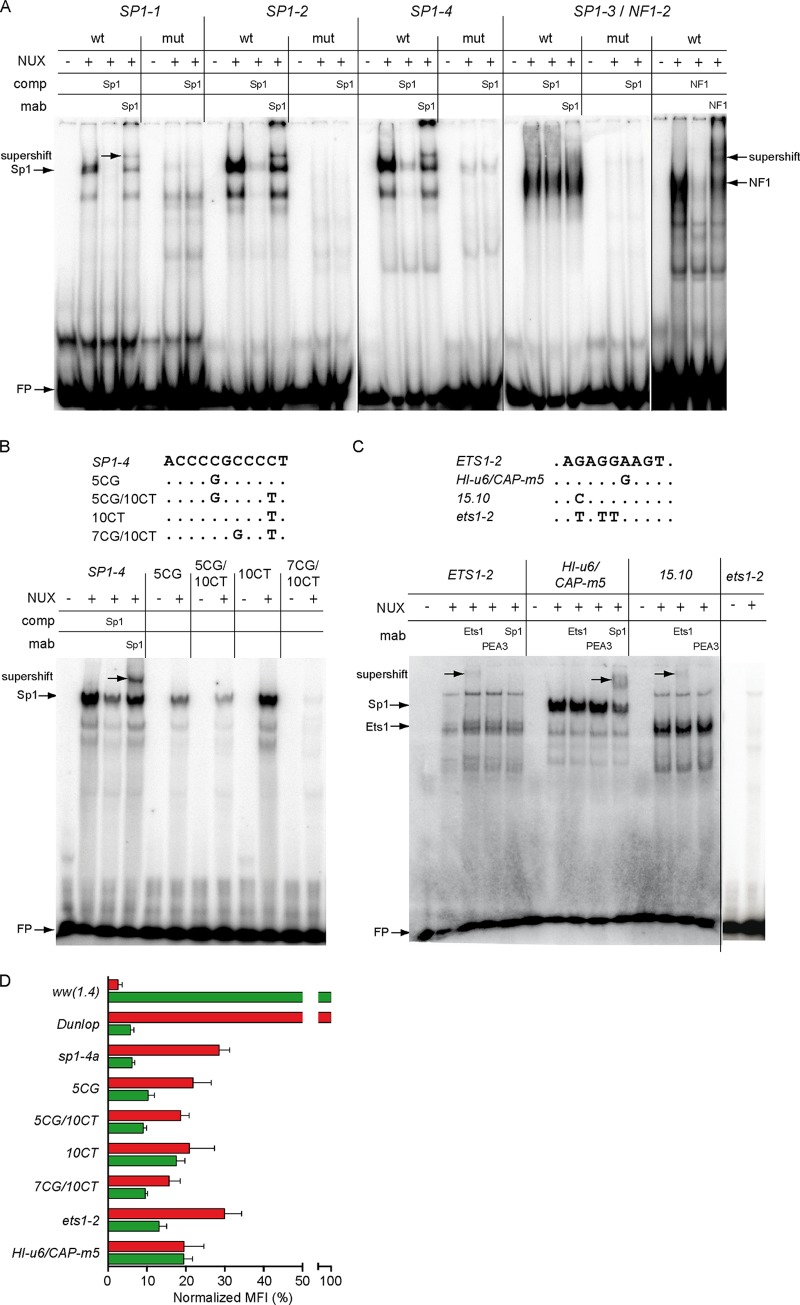
FIG 7.
Binding of Sp1 and Ets1 to archetype and mutant NCCR sequences and reporter gene expression. (A) Electrophoretic mobility shift assays (EMSAs) were performed using labeled and duplexed oligonucleotides carrying the indicated NCCR sequences with nuclear extracts (NUX) alone or in the presence of unlabeled competitor (comp) or monoclonal antibody (MAb), where indicated. Names of oligonucleotides (top row) refer to putative binding sites; wt, wild type sequence; mut, mutated binding site; NUX, 10 μg of RPTEC nuclear extract; comp, competition with 1 pmol unlabeled oligonucleotide with Sp1 or NF1 consensus sequence; MAb, 200 ng of monoclonal Sp1 or NF1 antibody for supershift; FP, free probe. (B) (Top) Alignment of naturally occurring SP1-4 mutants as described by Priftakis et al. (27). Names refer to the position relative to the SP1-4 binding site start and type of exchange. (Bottom) Binding analysis of SP1-4 mutants. (C) Oligonucleotides bearing the indicated archetype or mutant NCCR sequences were incubated with nuclear extracts as described in Materials and Methods. Archetype, ETS1-2 sequence from the BKPyV ww(1.4) strain; HI-u6/CAP-m5, sequence derived from a natural variant found in a patient with systemic BKPyV replication and capillary leak pathology (29); 15.10, sequence derived from a natural variant del(15.10) from kidney transplant patients with nephropathy (8); NUX, 10 μg of RPTEC nuclear extract; MAb, 200 ng monoclonal Ets1, PEA3, or Sp1 antibody for supershift; FP, free probe. (D) The indicated mutant NCCR bidirectional reporter constructs were transfected into HEK293 cells and grouped according to normalized MFI (Fig. 1D) (see Materials and Methods).