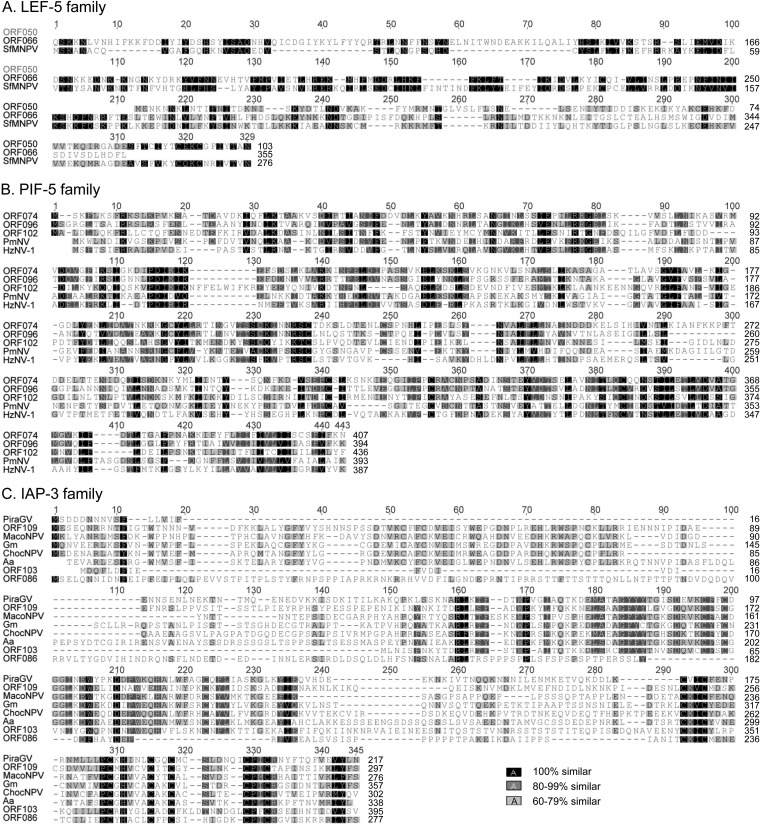
FIG 2.
Amino acid sequence alignments of ToNV LEF-5 (ORF050 and ORF066), PIF-5 (ORF074, ORF096, and ORF102), and IAP-3 (ORF086, ORF103, and ORF109) multigenic family members with viral or insect homologs. (A) LEF-5 family; SfMNPV, Spodoptera frugiperda MNPV (YP_001036358); (B) PIF-5 family; HzNV-1, Heliothis zea NV-1 (AAN04370), and PmNV, Penaeus monodon NV (YP_009051848); (C) IAP-3 family; PiraGV, Pieris rapae GV (AGS18838); MacoNPV, Mamestra configurata NPV (NP_613222); Gm, Galleria mellonella (ACV04797); ChocNPV, Choristoneura occidentalis NPV (YP_008378620); Aa, Aedes aegypti (EAT39096). Alignments were generated using the MAFFT alignment plugin under Geneious and then eventually manually refined. Each multiple alignment is presented with a global numbering located above the aligned sequences and a specific numbering located on the right side of each considered sequence. The similarity color code shown on the bottom right applies to all parts of the figure.