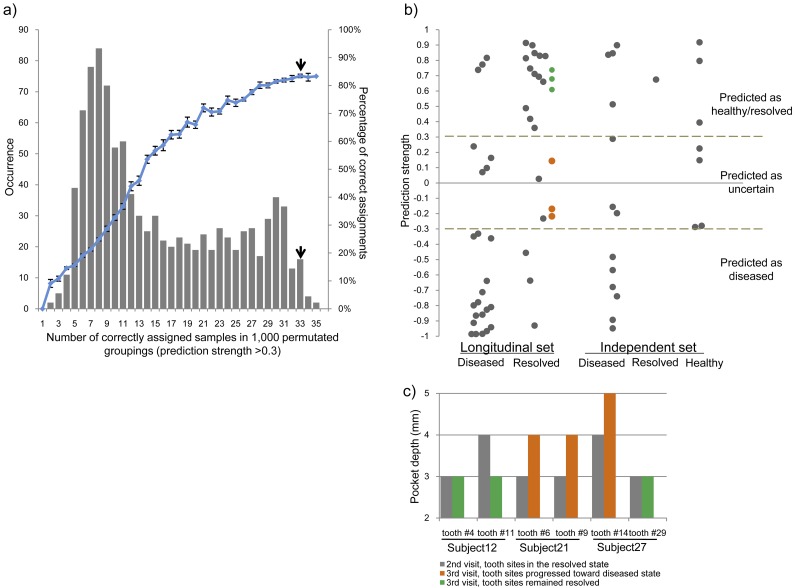
FIG 6 .
Classification of clinical states based on the subgingival microbiome profile. (a) Prediction accuracy using a weighted gene-voting algorithm and leave-one-out cross-validation. The number of correctly assigned samples based on the clinical grouping (indicated by the top short black arrow) was significantly higher than those in permutated groupings. The clinical grouping also had a significantly higher prediction accuracy (84.6%). (b) Classification of 48 longitudinal samples and an independent set of 21 samples based on the subgingival microbiome profile. The independent set consisted of 13 samples collected in the diseased state and one sample collected in the resolved state. Additionally, we included seven samples obtained from healthy subjects in the Human Microbiome Project (36). (c) Follow-up examination demonstrates our ability to predict disease progression. Six tooth sites in three patients were examined again during the third clinic visit. The directions of the clinical progressions of all six sites were correctly predicted based on the preceding microbiome profile at the second clinic visit. The tooth sites that remained healthy at the third visit were predicted as resolved (shown in green [Fig. 6b]), and the tooth sites that progressed toward or became diseased were predicted to be between the resolved and diseased states (shown in orange [Fig. 6b]).