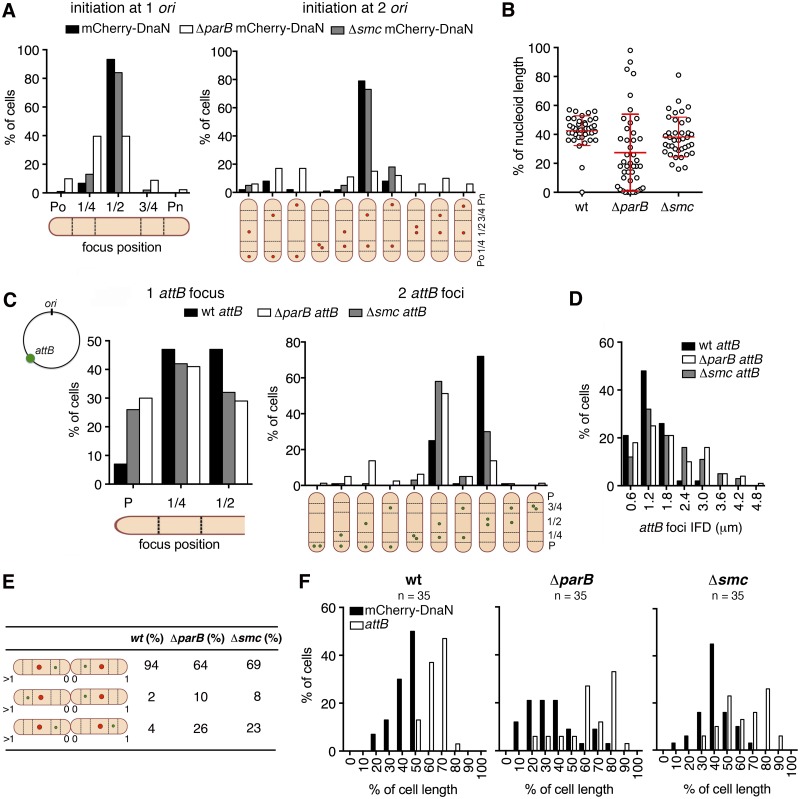
FIG 3 .
Subcellular localization of ori and replisome assembly require ParB but not SMC. (A) Distribution of positions of mCherry-DnaN foci relative to the old cell pole (Po) at initiation of DNA replication in wild-type (black bars), ΔparB (white bars), and Δsmc (grey bars) cells (n = 100 per strain) that initiate replication after cytokinesis (one ori, left panel) or before cytokinesis (two ori, right panel). Timing of cytokinesis is defined by the first appearance of Wag31-GFP at midcell. Cells that initiate DNA replication before cytokinesis are classified on the basis of the positions of the two mCherry-DnaN foci (schematic). (B) Positions of mCherry-DnaN foci at initiation of DNA replication relative to the SYTO green-stained nucleoid length in wild-type (wt), ΔparB, and Δsmc cells (n = 40 per strain). (C) Distribution of positions of FROS-attB foci in wild-type (wt; black bars, n = 135), ΔparB (white bars, n = 112), and Δsmc (grey bars, n = 196) cells containing one FROS-attB focus (left panel) or two FROS-attB foci (right panel). Cells with two FROS-attB foci are classified on the basis of the positions of the two foci (schematic). (D) Distribution of IFDs of FROS-attB foci in wild-type (wt; black bars, n = 305), ΔparB (white bars, n = 351), and Δsmc (grey bars, n = 261) cells with two FROS-attB foci. (E) Positions of mCherry-DnaN foci (red circle) and FROS-attB foci (green circle) at the time of replisome assembly in sibling cells. Three scenarios and their relative frequencies are shown for wild-type (wt), ΔparB, and Δsmc cells. (F) Distribution of positions of mCherry-DnaN foci (black bars) and FROS-attB foci (white bars) at the time of replisome assembly relative to the old cell pole (pole age, ≥1) in wild-type (wt), ΔparB, and Δsmc cells (n = 35 per strain).