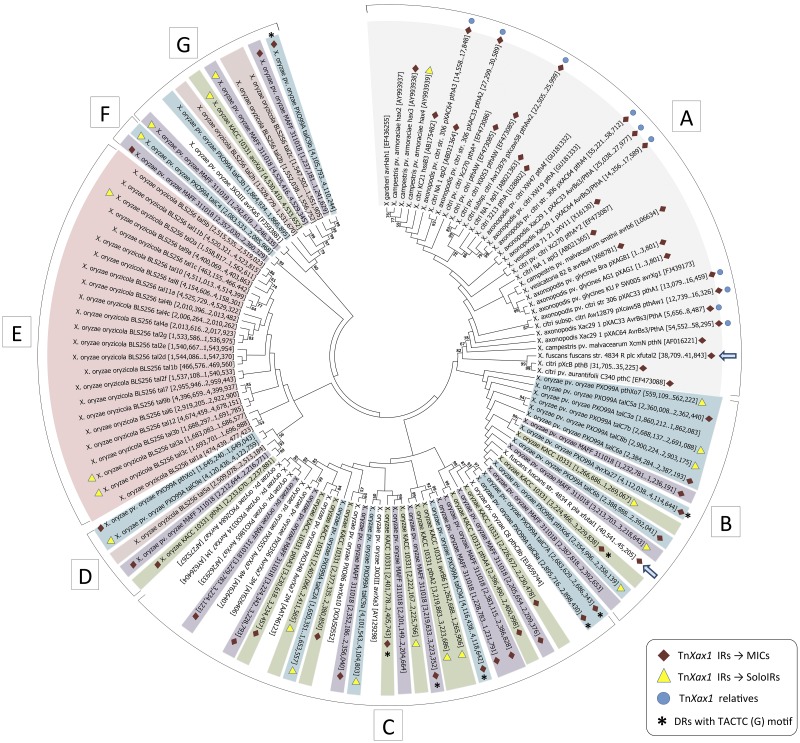
FIG 3 .
Molecular phylogenetic analysis by maximum-likelihood method of the TALEs genes found in completely sequenced Xanthomonas genomes. Each TALE gene is shown with its respective genomic coordinates (for TALEs extracted from complete genomes) or GenBank accession number inside the brackets. Symbols: red diamonds, TALE genes associated with MIC structures; yellow triangles, TALE genes associated with a solo IR flanking one of their extremities; blue circles, genomes which carry TnXax1 or related structures; asterisks, MICs with the direct TACTC(G) target repeat; blue arrow, the TALE gene from X. fuscans subsp. fuscans strain 4834-R plasmids pla and plc. TALEs from different X. oryzae pathovars are highlighted as follows: blue, X. oryzae pv. oryzae strain PXO99A; green, X. oryzae pv. oryzae strain KACC 10331; purple, X. oryzae pv. oryzae strain MAFF311018; pink, X. oryzae pv. oryzicola strain BLS256. TALEs from X. citri, X. axonopodis, X. campestris pv. vesicatoria, and X. campestris are highlighted in gray.