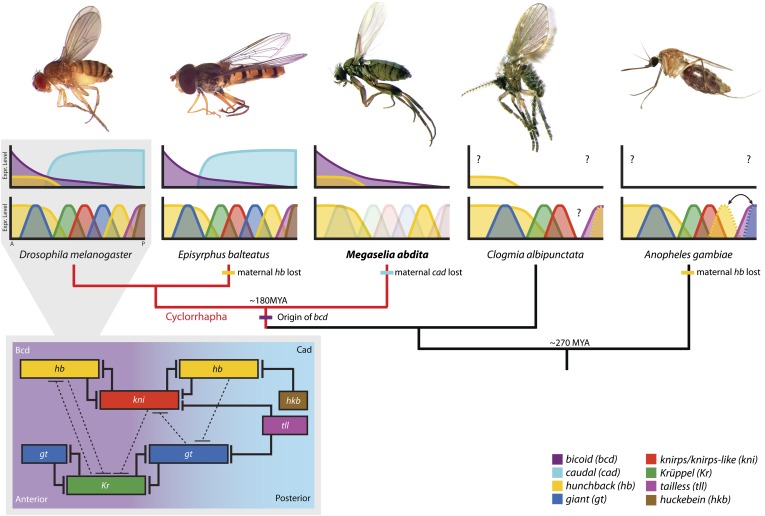
Figure 1. The evolution of the dipteran gap gene network.
A simplified phylogenetic tree of the order Diptera indicates the relative position of M. abdita with regard to other species of flies, midges, and mosquitoes in which gap genes have been studied in some detail. M. abdita belongs to the brachyceran infra-order Cyclorrhapha (marked in red); paraphyletic nematoceran lineages are shown in black. Only cyclorrhaphan flies have a bcd gene. Other maternal gradients have been lost along various branches of the tree (as indicated). For each species, we show an image of the adult (top), as well as a schematic representation of the spatial arrangement of maternal gradients (middle), and gap gene expression domains (bottom). Y-axes show expression levels (in normalised arbitrary units); X-axes show position along the major axis of the embryo (A: anterior, P: posterior). See key for colour coding. Solid colours indicate previously published expression patterns; faded colours represent previously unknown patterns reported in this study. Question marks indicate unknown maternal gradients or potentially missing gap domains. The inset (grey background) shows the gap gene network in D. melanogaster. Within the inset, background colour indicates major maternal regulatory inputs, boxes show the position of gap domains along the A–P axis; T-bars represent strong (solid) or weak (dashed) cross-repressive interactions among gap genes. Species (families): D. melanogaster (Drosophilidae): vinegar fly; E. balteatus (Syrphidae): marmalade hoverfly; M. abdita (Phoridae): scuttle fly; C. albipunctata (Psychodidae): moth midge; A. gambiae (Culicidae): malaria mosquito. All images are our own except A. gambiae image taken by Muhammad Mahdi Karim (source: Wikimedia Commons). See text for details.