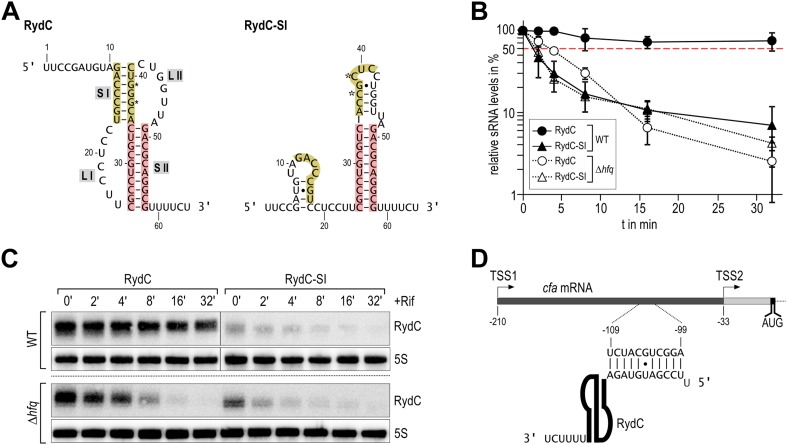
Figure 1. Association of RydC with the chaperone Hfq and its pseudoknot fold are required for sRNA stability.
(A) Secondary structure of the RydC pseudoknot and the predicted alternative structure are formed by the double mutant RydC-SI. Substitutions G37C and G39C are indicated with asterisks. (B, C) Stabilities of RydC and RydC-SI were determined by Northern blot analyses. Total RNA samples were extracted prior to and at indicated time-points after inhibition of transcription by rifampicin in late exponential phase (OD600 of 1) from Salmonella strains ΔrydC (JVS-0291) or Δhfq (JVS-0584), carrying plasmids expressing RydC (pKF42-1) or RydC-SI (pKF60-1) from the constitutive PL promoter. Error bars represent standard deviation calculated from three biological replicates. (D) Predicted duplex formed between the RydC 5′-end (nts 2–11) and the longer isoform cfa mRNA (nts −99 to −109 relative to the translational start site) originating at transcriptional start site 1 (TSS1).