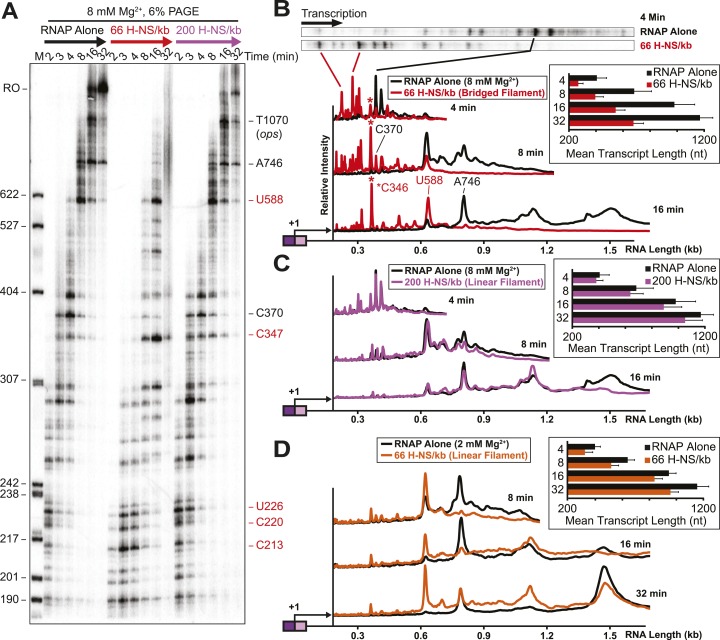
Figure 2. H-NS dramatically decreased transcript elongation in vitro.
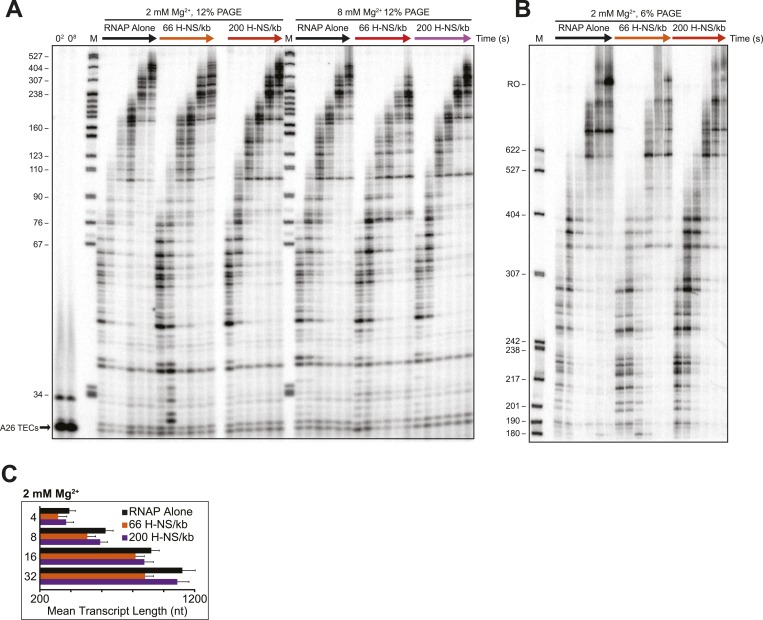
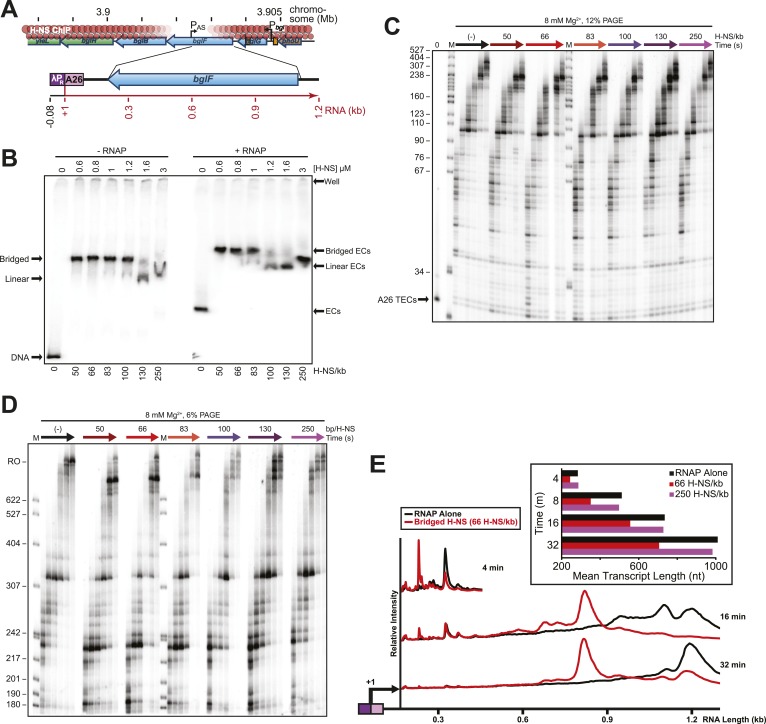
(A) In vitro transcription in the presence of 66 H-NS/kb or 200 H-NS/kb filaments at 20°C, 8 mM Mg2+, and 30 µM each NTP. ECs (10 nM) were formed at the end of the C-less cassette on the λPR-bgl template (A26 ECs) and then incubated with H-NS. Samples were removed at 2, 3, 4, 8, 16, and 32 min after addition of NTPs and separated by 6% PAGE. M, 5′ end-labeled, MspI-digested pBR322 marker. RO, run-off RNA. Pauses mapped to single-nt resolution in Figure 2—figure supplement 1 and Table 1 are indicated on the right side of the gel in red for H-NS-stimulated pauses and black for H-NS independent pauses. (B, C) Densitometry profiles of transcripts produced at 8 mM Mg2+ and 20°C from the λPR-bgl template in 66 H-NS/kb or 200 H-NS/kb filaments (B and C, respectively) or without H-NS (see ‘Materials and methods’). In (B), the 4-min time point from the gel shown in (A) is displayed horizontally to allow alignment with the densitometry profile (larger transcripts are to the right). Key pauses are marked in the profiles. Insets, mean transcript lengths and standard deviations were calculated from at least four independent experiments. (D) Densitometry profiles of transcripts produced at 2 mM Mg2+ and 20°C from the λPR-bgl template in 66 H-NS monomer/kb compared to without H-NS. Inset, mean transcript lengths and standard deviations were calculated from at least four independent experiments.