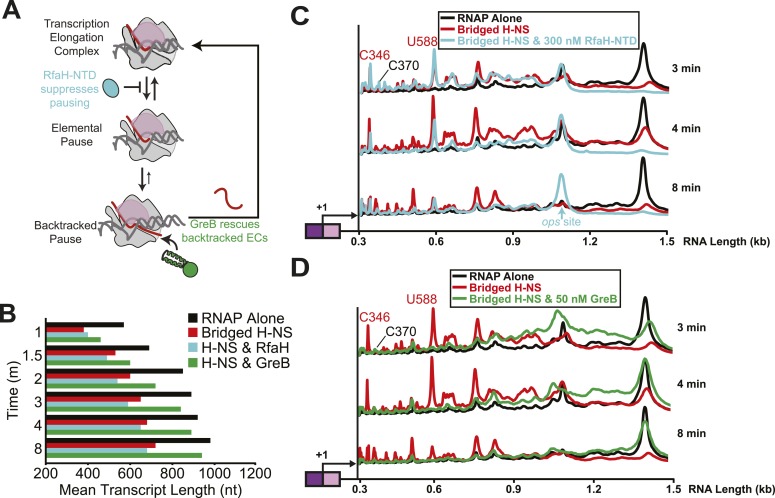
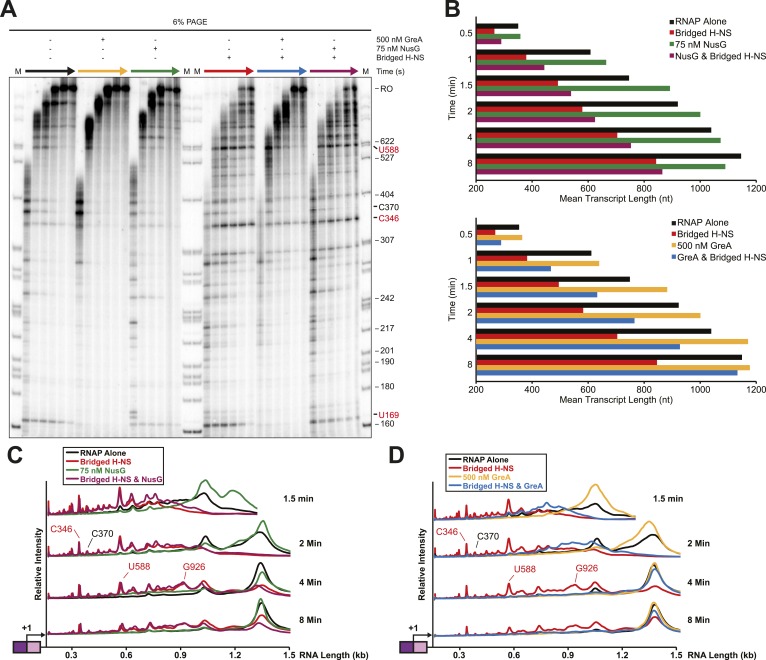
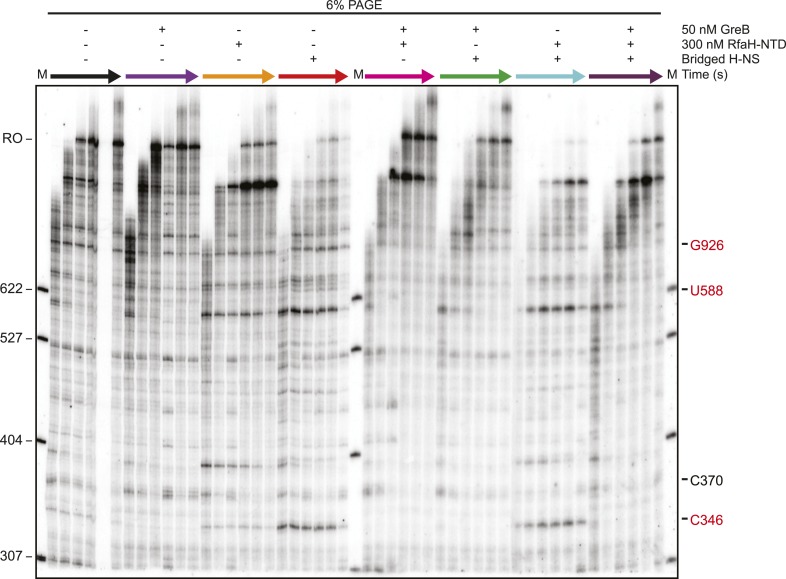
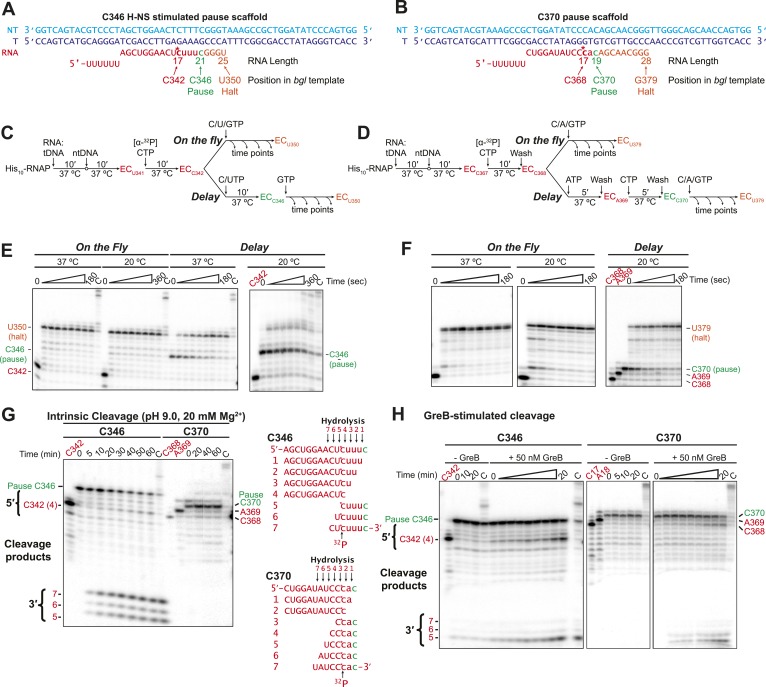
Figure 6. Bridged H-NS filaments induced RNAP backtracking, which was rescued by GreB.
(A) Steps in pausing affected by the NusG-like N-terminal domain (NGN) of RfaH (RfaH-NTD) or Gre factors (e.g., GreB). Binding of the NGN RfaH-NTD (cyan) to the clamp domain (pink) of RNAP inhibits clamp motion and suppresses entry into pause states (Sevostyanova et al., 2011). The duration of pausing once paused ECs form can be increased by backtracking of DNA and RNA through RNAP, during which the 3′ RNA enters the RNAP secondary channel. GreB promotes endonucleolytic cleavage of the backtracked RNA in the RNAP active site to convert an offline paused EC back to an active EC (Laptenko et al., 2003). (B) Mean transcript lengths were averaged from two independent experiments. (C, D) Densitometry profiles of transcripts produced at 20°C, 12 mM Mg2+, and 1 mM each NTP from the λPR-bgl template in bridged filaments (66 H-NS/kb) with or without 300 nM RfaH-NTD (C) or 50 nM GreB (D). Samples were removed at 0.33, 0.66, 1, 1.5, 2, 3, 4, 8, and 16 min after addition of NTPs and separated by denaturing PAGE.