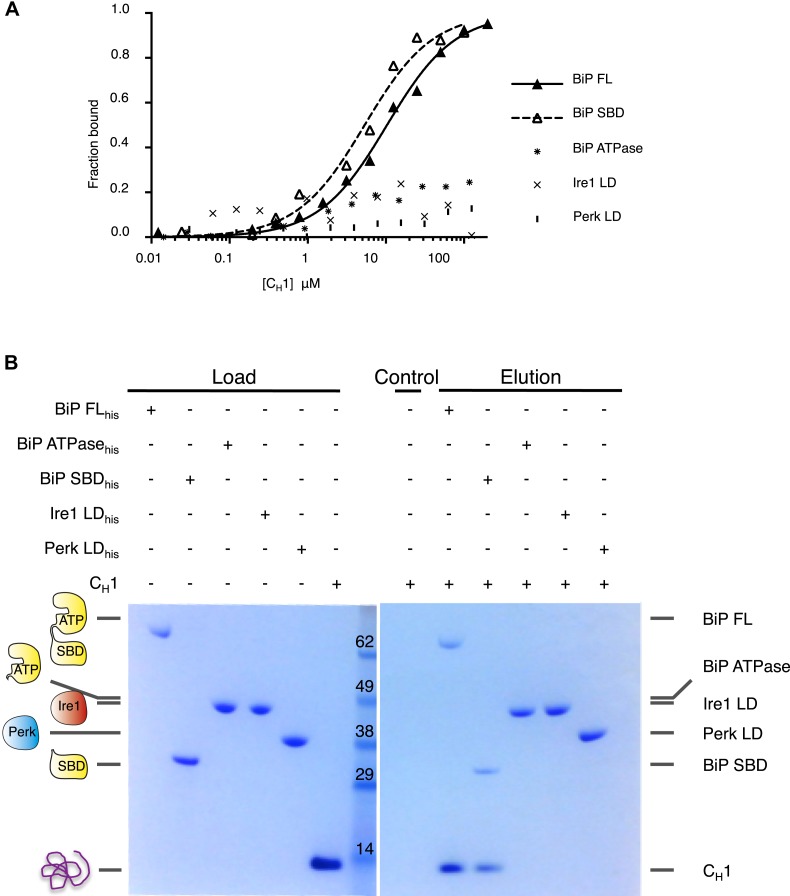
Figure 4. Unfolded protein CH1 binds to canonical BiP substrate binding domain without binding to UPR luminal domains.
(A) MST binding curves for CH1 binding to full-length BiP (Kd = 8.7 μM), BiP's ATPase domain (no binding), BiP's substrate binding domain (Kd = 5.1 μM), Ire1 luminal domain (no binding) and Perk luminal domain (no binding). (B) Pull down experiment showing CH1 binding to both full-length and substrate binding domain of BiP only, with no interaction observed to luminal domains, reaffirming the data for CH1 interactions using MST in part A.