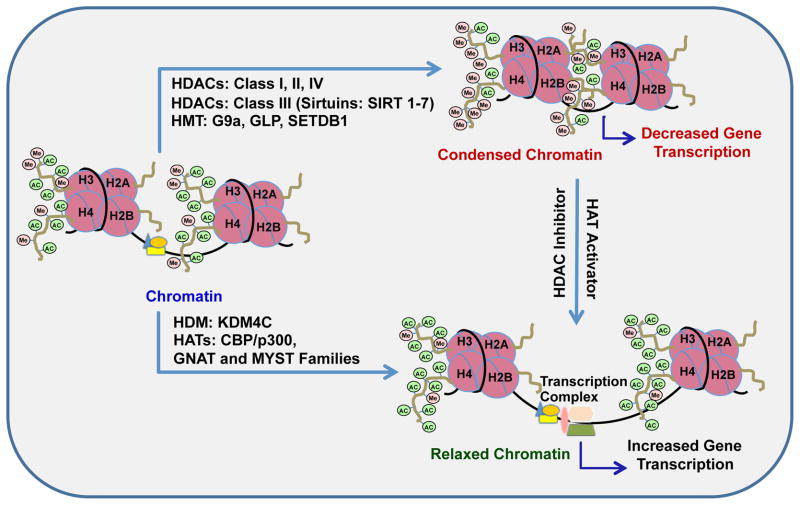
Figure 1.
Schematic representation of the epigenetic mechanisms of acetylation and methylation of lysine (K) residues at histone H3 and H4 tails that mediate the switching between ‘open’ (relaxed) and ‘closed’ (condensed) chromatin structures. Acetylation of histone H3 at K9, K14, K18 and K56 and of H4 at K5, K8, K12 and K16 are believed to relax the chromatin structure making it more accessible to transcription factors and chromatin remodelers, thereby promoting transcription, whereas methylation at these lysine residues condenses the chromatin followed by reduced gene transcription. Histone deacetylases (HDACs) remove acetyl groups and histone methyltransferases (HMTs) attach methyl groups at lysine residues. HDACs are classified into four different classes (class I–IV), of which the class III, sirtuins, are NAD+ dependent and comprise of seven different isoforms (SIRT 1–7). HDACs classes I (HDAC 1, 2, 3, and 8), II (HDAC 4–7, 9, and 10) and IV (HDAC11) are Zn+ dependent enzymes. Histone methyltransferases such as G9a, GLP and SETDB1 can catalyze the methylation of H3-K9 contributing to the restrictive state of the chromatin. Histone demethylases (HDMs) (e.g. KDM family) remove the methyl groups from lysine residues associated with repressive marks (e.g. H3K9) in the tails of histones facilitating the relaxation of chromatin. Histone acetyltransferases (HATs) such as GNAT and MYST families of enzymes and CREB-binding protein (CBP)-p300 complex transfer acetyl groups leading to relaxed chromatin (Allis et al., 2007; Kouzarides, 2007).