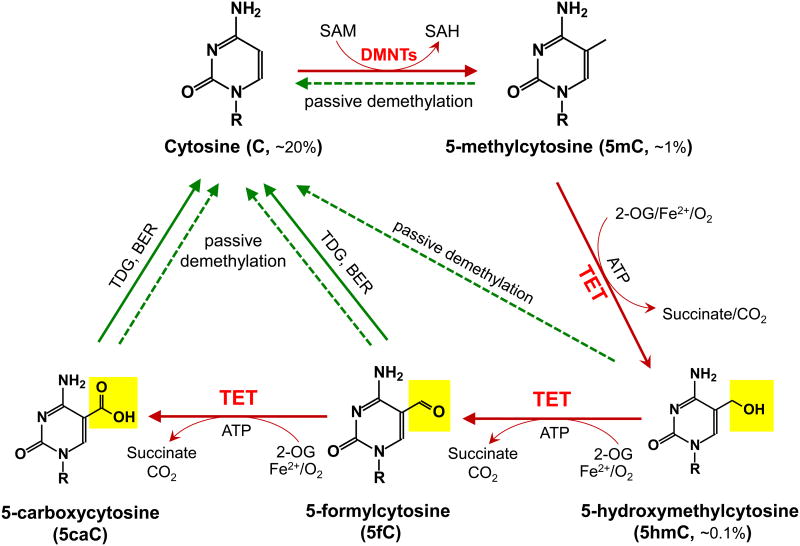
Figure 1. Schematic of major DNA methylation and demethylation pathways in mammals.
DNA methylation occurs almost exclusively as symmetrical methylation at the carbon-5 position of cytosine in the context of the dinucleotide CpG. DNMTs methylate cytosine (C, ∼20% of all bases) to yield 5-methylcytosine (5mC, ∼1% of all bases and ∼60% of all CpGs) by transferring the methyl group from S-adenosylmethionine (SAM) to cytosine. TET enzymes oxidize 5mC to 5-hydroxymethylcytosine (5hmC, ∼0.1% of all bases), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC) (together, oxi-mC). Through oxi-mC production, TET proteins mediate multiple pathways of DNA demethylation, including thymine DNA glycosylase (TDG)-mediated base excision repair (BER) of 5fC:G and 5caC:G basepairs and replication-dependent passive demethylation. A recent study showed that TET proteins can oxidize thymine to 5-hydroxymethyluracil (5hmU) [143], and other studies have suggested that Activation-induced deaminase (AID)/APOBEC can mediate the deamination of 5hmC to 5hmU followed by TDG-mediated BER (reviewed in [11]). Although 5hmU:G mismatches can also be excised by TDG [144], these pathways are less well-characterized and so are not depicted here.