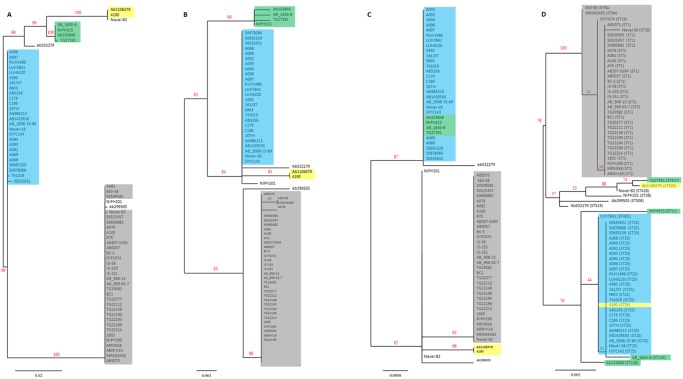
Fig 3. Phylogenetic trees of cas1, csy1, csy4 and the concatenated MLST sequences.
The phylogenetic trees were based on aligned nucleotide sequences of (A) 920 bp of cas1 from 69 isolates of Acinetobacter baumannii, (B) 1251 bp of csy-1 from 68 isolates of A. baumannii since csy1 was lacking in isolate Naval-82, (C) 615 bp of csy4 from 69 isolates of A. baumannii, and (D) 2976 bp of concatenated MLST sequences of 69 isolates of A. baumannii. MUSCLE, Gblocks, PhyML, and TreeDyn were used for nucleotide alignment and tree construction. One hundred bootstraps were used for bootstrap analysis. Branch support values were displayed in %. Isolates of the CRISPR-cas subtype I-Fb pathways of evolution 1, 2, 3, and 4 were highlighted in gray, blue, green, and yellow, respectively. The corresponding sequence type (ST) was presented next to the name of each isolate in (D).