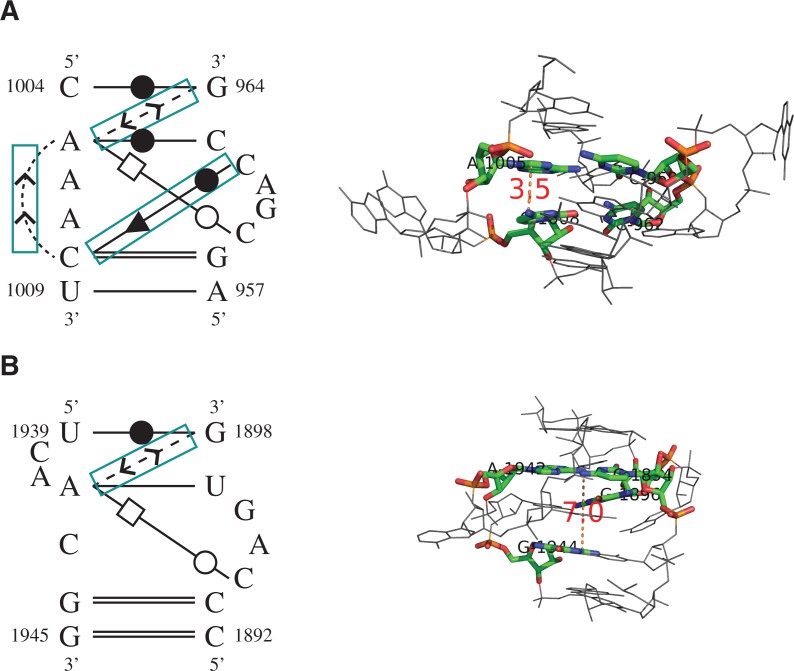
FIGURE 6.
The base-interaction patterns (left panel) and 3D structures (right panel) of (A) a true C-loop motif instance and (B) an unrelated motif instance. RNAMotifScan ranks A below B; however, RNAMotifScanX is capable of correctly ranking A above B. RNAMotifScanX corrects the ranking by considering the stacking interactions and the crossing base pair. In the left panel, conserved base interactions that are uniquely aligned by RNAMotifScanX are highlighted using the cyan boxes. The right panel shows that the lack of these interactions in B leads to significant structural changes of the core nucleotides (colored, with distance measures to highlight the variation between the same-strand stacked nucleotides, i.e., A1005, C1008 in A and A1942, G1944 in B) as well as the entire structure.