FIGURE 7.
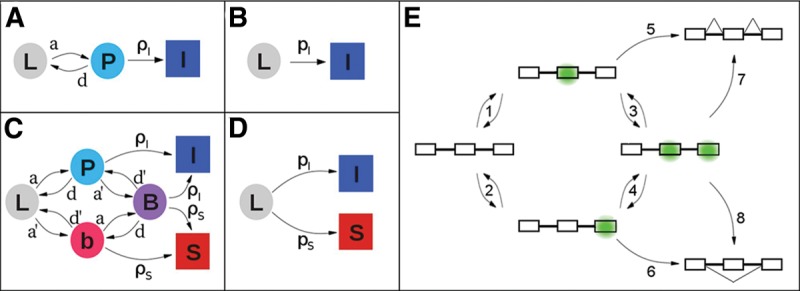
Complex kinetics can be described in simpler terms. The squares and circles represent different states of a pre-mRNA molecule: (L) “naked” transcript; (P) exon of interest in an exon definition complex (EDC) with the downstream exon either not present or present but not in an EDC; (b) downstream exon in an EDC with the exon of interest not in an EDC; (B) both exons in EDCs; (I) (inclusion) and (S) (skipping) represent molecules that have either committed to or achieved their respective splicing outcomes. The arrows represent transitions between states, and are labeled with rate constants: (a) and (d) association and dissociation, respectively, of the complex on the exon of interest; (a′) and (d′) the same for the downstream exon; (ρI) and (ρS) commitment to inclusion and skipping, respectively, of the exon of interest. (A) Model for the splicing reactions before time τ. Importantly, the transition from P to I is independent of the presence of exon 3. (B) Simplified model before time τ; pI amalgamates a, d, and ρI. (C) Model for the splicing reactions after time τ. (D) Model after time τ simplified analogously to B. pS amalgamates a′, d′, and ρS. See Supplemental Material for details. (E) Cartoon showing the states implied in C for a pre-mRNA molecule depicting EDCs (green). Steps 1–4 represent the formation or loss of EDCs; Steps 5–8 represent commitments to the splicing outcome shown.
