Fig. 2.
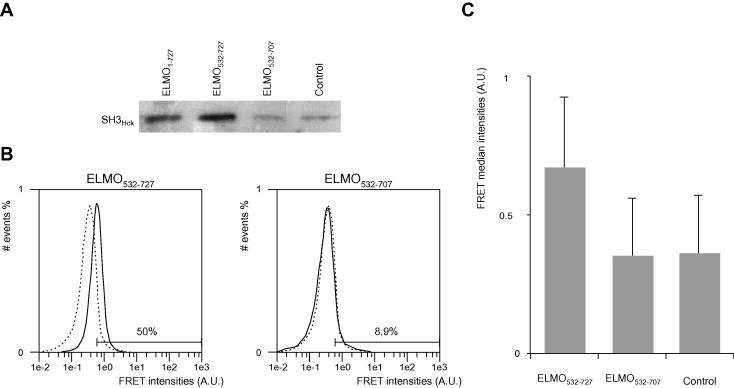
Binding of ELMO1 domains to SH3 domain of Hck in the cellular context. (A) SH3Hck binding to ELMO1 is dependent of ELMO1 polyproline motif in co-transfected HEK293T cell extract: CLIP-SH3Hck was expressed either alone or with SNAP-ELMOWT and its various deletion mutants (ELMO532–727 or ELMO532–707) in HEK293T cells. Cell lysates were then incubated with SNAP-capture magnetic beads. Proteins were eluted after TEV digestion, separated by SDS–PAGE and the presence of retained CLIP-SH3Hck by western-blot using a polyclonal antibody anti-SNAP-tag (which is also specific for CLIP tag). The control well (cells mono-transfected with CLIP-SH3Hck) shows the residual non-specific signal. (B) Flow cytometry distribution of FRET intensities of HEK293T cells co-transfected with SH3Hck and ELMO1 deletion mutants: Superposition of the histograms of the FRET intensities (acceptor fluorescence) from the control cells (transfected with CLIP-SH3Hck, dash line) and cells transfected with CLIP-SH3Hck and either with SNAP-ELMO532–727 or SNAP-ELMO532–707 (plain lines). The FRET signal was measured in the V2 channel Ex 405 nm, Em 525 ± 25 nm. A shift of the maximum of the double positive population is observed for the measured FRET channel for SNAP-ELMO532–727 relative to the negative control, while no difference is noticeable for SNAP-ELMO532–707. The shift is also expressed as the ratio of cells above the threshold in comparison with the total number of cells in the gated region (donor positive cells, see Section 2). The data shown originate from a single experiment representative of three independent experiments. (C) Bar graph of FRET median values: FRET intensities median values of the histograms presented in B. The standard deviation errors of the donor positive population are shown.
