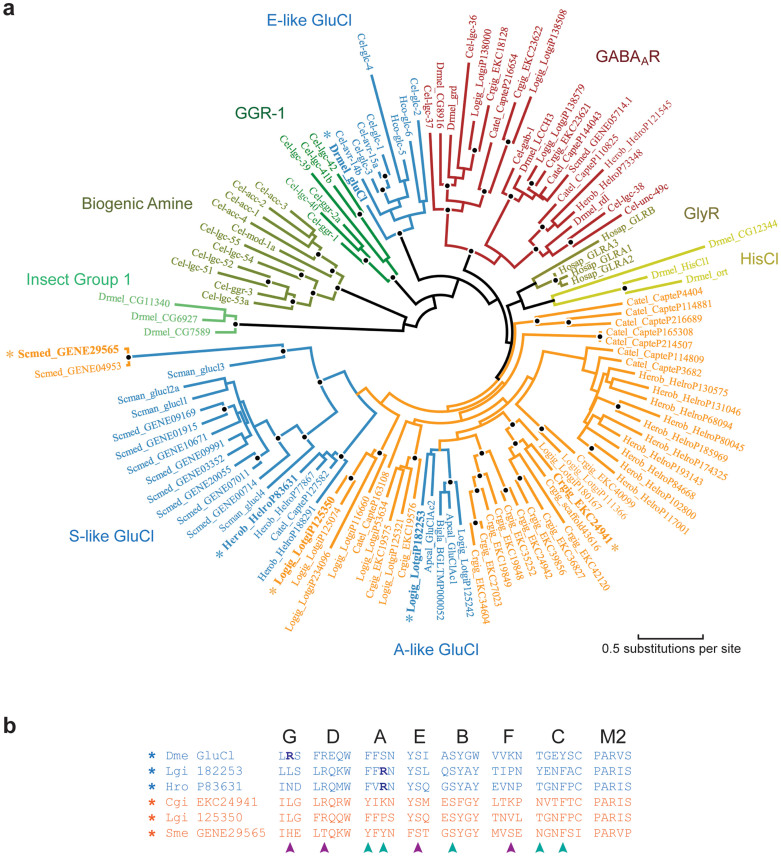
Figure 4. GluCl phylogeny.
(a) Maximum likelihood phylogeny of anionic pLGIC subunit protein sequences representing the major clades identified from vertebrates and Ecdysozoa2, S. mansoni and A. californica GluCls, and newly identified, predicted anion channel subunits from the molluscs C. gigas and L. gigantea, the platyhelminth S. mediterranea and the annelida C. teleta and H. robusta. Major clades are identified in different colors and labeled, with sequences containing the glutamate-binding motif colored blue. Branch lengths are proportional to the number of substitutions per amino acid, and nodes with at least 90% SH and at least 70% bootstrap support are indicated by a black circle. Gene identifiers are from GenBank, and nematode gene names follow Beech et al.52. (b) Partial amino acid sequence alignment of selected subunits from (a). AAA motif and arginine residues are indicated by arrows as in Fig. 1b (cyan, principal face; magenta, complementary face). Note how orange sequences differ from verified GluCl subunits, in the absence of Loop G or A arginine residues, the presence of the initial Loop B glutamate residue, and/or the absence of the initial Loop C threonine residue. Dme, Drosophila melanogaster; Lgi, Lottia gigantea; Hro, Helobdella robusta; Cgi, Crassostrea gigas; Sme, Schmidtea mediterranea.