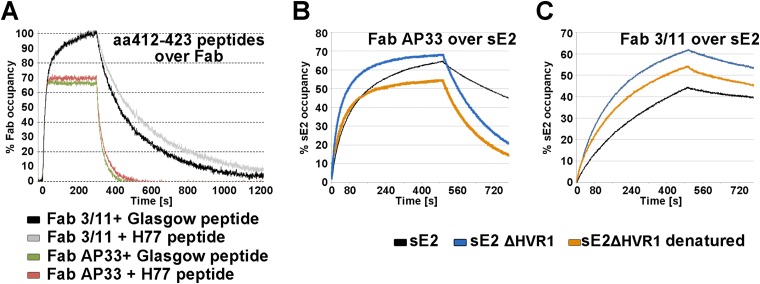
FIG 5.
SPR analysis of Fab binding to aa 412 to 423. To facilitate direct comparisons of the kinetics of binding to different Fab fragments, the binding response was normalized in each case, as described in Materials and Methods, and expressed as the occupancy of binding sites. (A) Real-time SPR analysis of the interaction of the epitope peptides derived from genotype 1a strain Glasgow and H77 with immobilized Fab fragments. The respective epitope peptide was injected over a surface of covalently immobilized Fab at a flow rate of 50 μl/ml, and the binding response in resonance units (RU) was recorded as a function of time. To compensate for the small signal (due to the low molecular mass of the peptide ligand of ∼2 kDa), three independent experiments were performed, and the mean values with standard errors are presented in Table 3. The representative association/dissociation time course profile shown corresponds to an injection of 125 nM peptide over each of the Fabs, illustrating the different kinetic properties of the two Fab-peptide complexes. (B and C) Real-time SPR analysis of the binding of Fabs AP33 (B) and 3/11 (C) to the immobilized HCV E2 ectodomain. Fabs were injected over HCV sE2 immobilized by using an anti-Strep-Tag antibody at a flow rate of 30 μl/ml, and the binding response in RU was recorded as a function of time. Representative association/dissociation time course profiles correspond to the injection of Fabs (355 nM) over full-length immobilized HCV sE2 or native and denatured HCV sE2 ΔHVR1.