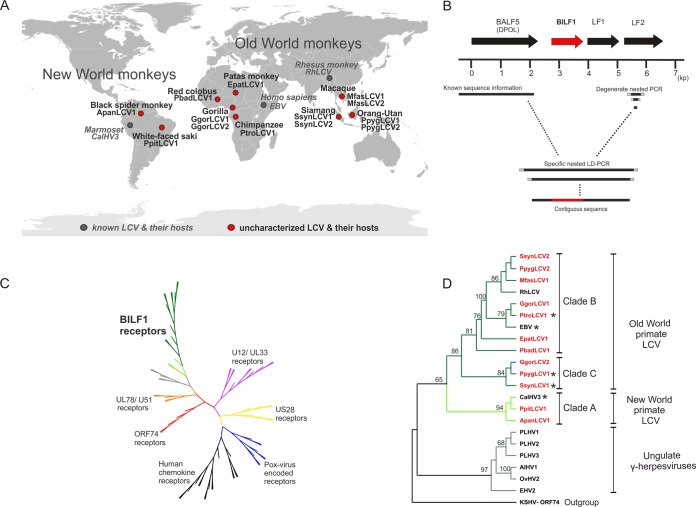
FIG 1.
Identification of novel BILF1 receptor sequences from uncharacterized LCVs of nonhuman primates and phylogenetic studies. (A) World map of primate hosts harboring novel and known LCVs. The viruses are indicated in red and gray circles (novel and known LCVs, respectively); names of viruses and their hosts are written in bold blac (novel viruses) and italic gray (known viruses). Three ape hosts (gorilla, siamang, and orangutan) harbor 2 distinct LCVs. (B) Map of amplified genes and diagram of PCR strategy. EBV ORF BALF5 (DPOL), BILF1, LF1, and LF2 are symbolized as arrows (BILF1 ORF in red). A scale (in kb) oriented on the EBV genome is given below. The right side shows degenerate nested primers (light gray squares) used to amplify part of the LF2 gene. A short solid black line represents the amplified fragment. The left side represents published sequence information for the DNA polymerase gene of the investigated LCV (solid black line). Based on both DPOL and LF2 sequences, specific primers (gray squares) were selected and long-distance PCR was performed. A final contiguous sequence of approximately 3.5 kb was obtained, including the BILF1 ORF (red). (C) Phylogenetic tree of viral and endogenous 7TM receptors. The amino acid sequences were aligned with MAFFT and an unrooted tree was constructed using the neighbor-joining algorithm. The published U12/UL33 (purple), US28 (yellow), poxvirus-encoded receptor (blue), human chemokine receptor (black), ORF74 (red), and UL78/U51 (orange branches) sequences were aligned with the published and newly detected BILF1 sequences (green branches) and subjected to phylogenetic analyses. (D) Phylogenetic tree of BILF1 receptor family. BILF1 amino acid sequences were aligned with MAFFT and a rooted tree was constructed, using the phyml/bootstrap algorithm and the KSHV ORF74 sequence as an outgroup. The BILF1 sequences of New World primate LCVs (clade A; bright green branches) separate from those of Old World primate LCVs (clades B and C; dark green branches). The BILF1-homologous sequences from gammaherpesviruses of ungulates (gray branches) cluster separately from the primate BILF1 sequences. Newly discovered BILF1 sequences are highlighted in red, and BILF1 receptors used for functional studies are marked with a star.