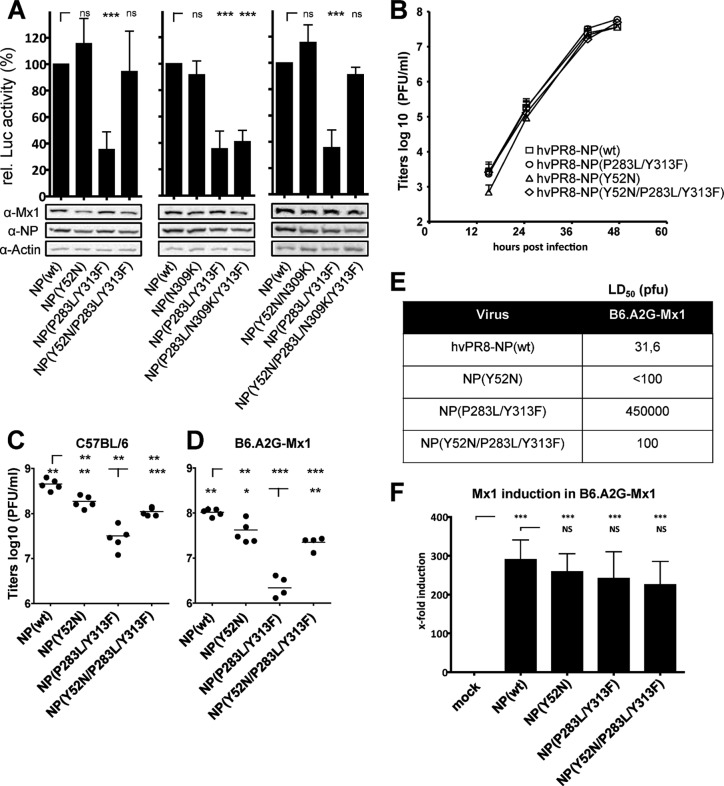
FIG 3.
Mx sensitivity of NP mutants at positions 52 and 309. (A) A polymerase reconstitution assay was performed as described in the legend of Fig. 1B, including the compensatory NP mutants. Relative polymerase activity was calculated as the ratio of luciferase activity in the presence of wild-type Mx1 compared to the luciferase activity in the presence of inactive Mx1(K49A). The activity with wild-type NP was set to 100%. Bars represent mean values with standard deviations of two independent experiments performed in triplicates. Two-way analysis of variance was performed to calculate P values. ns, not significant; ***, P < 0.001. Western blot analysis shows the expression levels of Mx1, NP, and β-actin in the cell lysates. (B) Growth kinetics of the recombinant hvPR/8 viruses on Calu-3 cells infected at an MOI of 0.001 in the presence of 0.5 μg/ml trypsin. Error bars indicate the standard deviations of one experiment performed in duplicates. (C and D) Lung titers of hvPR/8 viruses in C57BL/6 (C) and B6.A2G-Mx1 (D) mice upon intranasal infection with 200 PFU. At 48 h postinfection virus titers were determined in lung homogenates. Student's t test was performed to calculate P values. (E) MLD50 values of hvPR/8 encoding wild-type NP or the indicated NP mutants determined in B6.A2G-Mx1 mice (n = 5/group). (F) Detection of Mx1 expression by quantitative RT-PCR. Lung homogenates of B6.A2G-Mx1 mice (n = 5) infected with 200 PFU for 48 h were used to extract RNA and determine the expression of Mx1 and GAPDH by qRT-PCR. The Mx1 signals were normalized to the GAPDH levels. qRT-PCR results were calculated as Mx1-to-GAPDH ratio by the 2ΔCT method (59). The relative induction of Mx1 in mock-infected animals was set to 1. Bars represent mean values with standard deviations using data from five animals each (three animals in the mock control). Two-way analysis of variance was performed to calculate P values. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. α, anti.