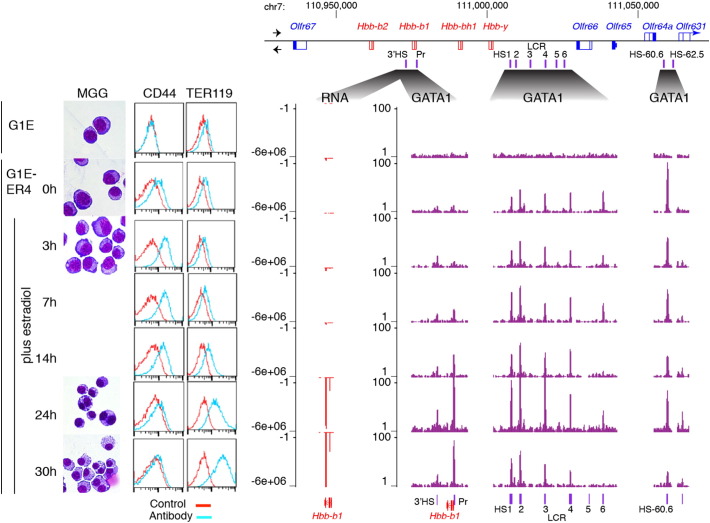
Fig. 1.
Illustrative changes in morphology, cell surface markers, RNA, and GATA1 occupancy during GATA1-dependent erythroid maturation. G1E cells and G1E-ER4 cells either untreated (0 h) or treated with E2 for increasing lengths of time (top to bottom) were examined by (left to right) microscopy after staining cytospins with May–Grunwald–Giemsa (MGG), and FACS after fluorescent staining with antibody against CD44 and TER119. Results of RNA-seq on polyA + RNA and ChIP-seq with antibody against GATA1 are shown on the right for the locus encoding beta-globins and olfactory receptors, specifically a 145,400 bp region at chr7:110,928,295-111,073,694 (NCBI37/mm9 assembly of the mouse genome). In the locus map, genes transcribed from left to right are above the line and those in opposite orientation are below the line. RNA-seq reads map to bottom (minus) strand of the assembly, and thus the quantitation of the RNA is plotted as negative numbers. The locus control region (LCR) is a complex enhancer regulating the Hbb genes encoding beta-globins. Additional DNase hypersensitive sites (HSs) even more distal from the Hbb genes are also shown.