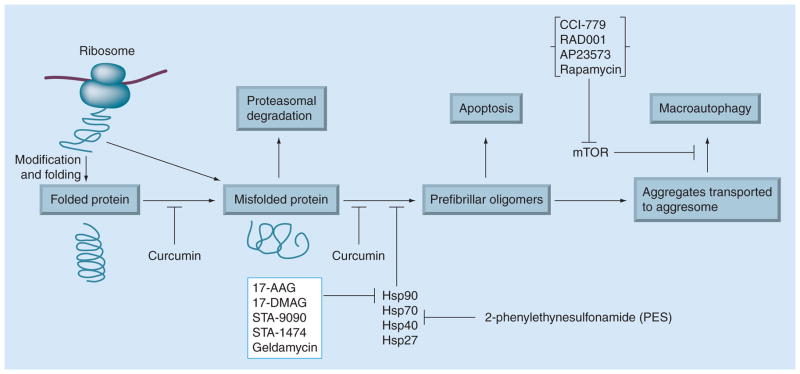
Figure 1. Model of protein misfolding and therapeutic targets in cancer.
Proteins are synthesized on ribosomes from the genetic information encoded in cellular DNA and undergo the major part of their folding in the cytoplasm after release from the ribosome. Newly synthesized proteins are translocated into the endoplasmic reticulum, where they fold into their 3D structures. Correctly folded proteins are transported to the Golgi complex and then delivered to the extracellular environment. Under certain circumstances, a folded protein is converted to a misfolded protein by various stresses, gene mutations or proteolysis. During normal conditions, misfolded proteins are detected by a quality-control mechanism and are degraded by the ubiquitin–proteosome pathway. A misfolded protein can be induced to form prefibrillar oligomers at higher concentrations and transported to form aggresomes. These aggregated proteins at the aggresome are targeted for degradation via macroautophagy. Accumulation of prefibrillar oligomers at the aggresome pathway may induce apoptosis. Curcumin, a potential antioxidant or anticancer compound may inhibit protein misfolding and/or aggregation. Protein aggregation can also be blocked by upregulation of molecular chaperones with heat shock protein (HSP)90 inhibitors (17-allylamino-demethoxygeldanamycin (AAG), 17-[dimethylaminoethylamino]-17-demethoxygeldanamycin (DMAG), STA-9090, STA-1474 and geldamycin) or HSP70 inhibitors, such as 2-phenylethyneusulfonamide. Aggregate clearance can be upregulated via inhibition of mTOR by rapamycin that inhibits macroautophagy. Analogues of rapamycin, such as CCI-779 (temsirolimus), RAD001 (everolimus) and AP23573, are likely to be the first mTOR-perturbing molecules to be approved for anticancer use in humans. Redrawn with permission of Cambridge University Press from [106].