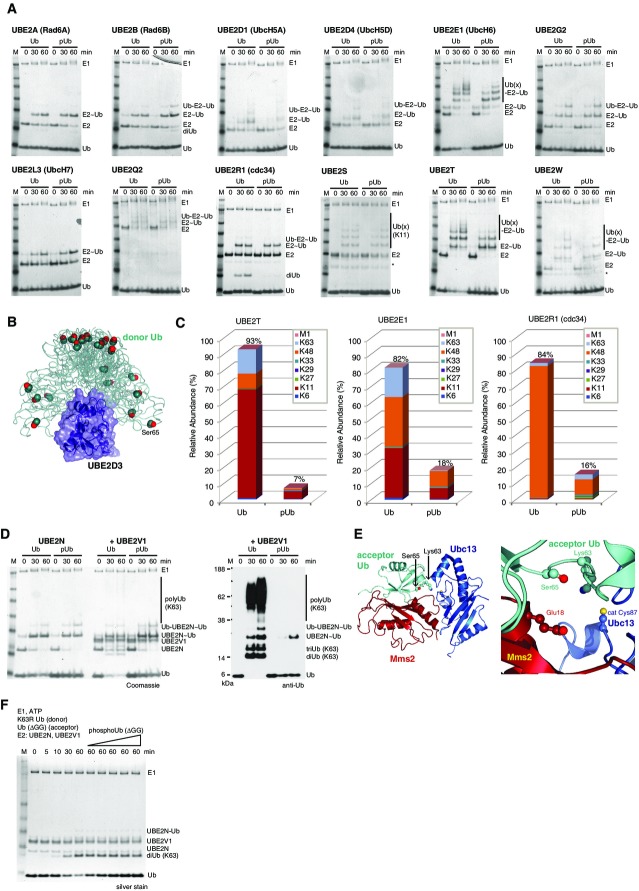
Figure 5.
Impact of Ub phosphorylation on E1 and E2 enzymes
- E1-mediated charging of E2 enzymes by Ub and phosphoUb in a time-course analysis. Non-reducing Coomassie-stained 4–12% SDS–PAGE gradient gels are shown, and bands are labeled. “˜Ub” refers to generation of a thioester, while “Ub-“refers to additional covalent ubiquitination of the E2 enzyme. * contaminant from E2 purification.
- SAXS-derived ensemble of UBE2D3 (Pruneda et al, 2011), where the E2 is shown under a blue surface and the Ub is shown as a green ribbon. Ser65 is shown in sphere representation. Ser65 does not contact the E2 enzyme in any orientation of the Ub at the E2 enzyme. PDB files were downloaded from the Klevit-lab website (http://depts.washington.edu/klvtlab/).
- Quantitative mass spectrometry analysis for UBE2T, UBE2E1 and UBE2R1 with Ub and phosphoUb using Ub AQUA peptides reveals impaired chain formation with phosphoUb (see also Supplementary Fig S10).
- Charging of UBE2N, and UBE2N/UBE2V1-mediated Lys63 chain assembly with Ub and phosphoUb. Charging of UBE2N as in (A) proceeds identically with both Ub and phosphoUb (left 6 lanes), while UBE2V1-mediated Lys63 chain assembly only proceeds with Ub but not phosphoUb (right 6 lanes and Western blot).
- Structure of Saccharomyces cerevisiae Ubc13-Mms2 (blue/red, Mms2 is a homolog of UBE2V1) with the acceptor Ub (cyan) bound non-covalently to Mms2 [pdb-id 2gmi (Eddins et al, 2006)]. Ser65 of the acceptor Ub interacts with Mms2, and Ser65 phosphorylation would prevent binding due to Mms2 Glu18.
- PhosphoUb competition assays for UBE2N/UBE2V1-mediated Lys63 diUb formation. Time course with K63R Ub (donor) and Ub ΔGG (acceptor) generates Lys63 diUb and is not inhibited upon increasing concentrations of phosphoUb, revealing phosphoUb does not compete with Ub for UBE2N/UBE2V1-mediated Lys63 diUb formation.