Figure 5.
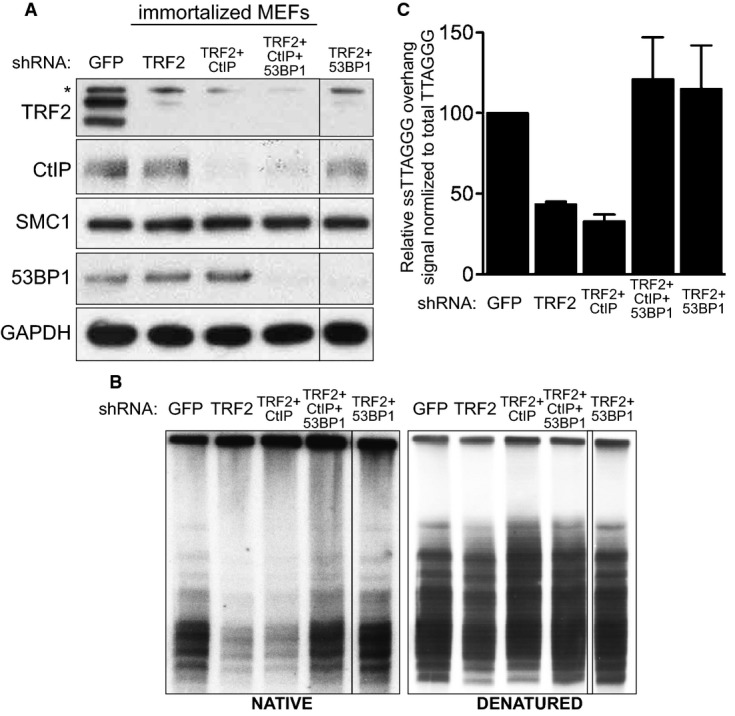
- Immortalized MEFs were infected with retroviruses expressing the indicated shRNAs, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. SMC1 and GAPDH were used as loading controls. *non-specific band.
- MboI- and AluI-digested DNA from cells treated as in (A) was resolved by pulsed-field gel electrophoresis and probed with end-labelled (AACCCT)4 probe. Representative pulsed-field gel samples run under native and denatured conditions are shown.
- Quantification of the 3′ overhang in cells treated as in (B). For each sample, the ss/total DNA ratios were expressed relative to the GFP shRNA-treated control. Error bars represent SD of two independent experiments.
