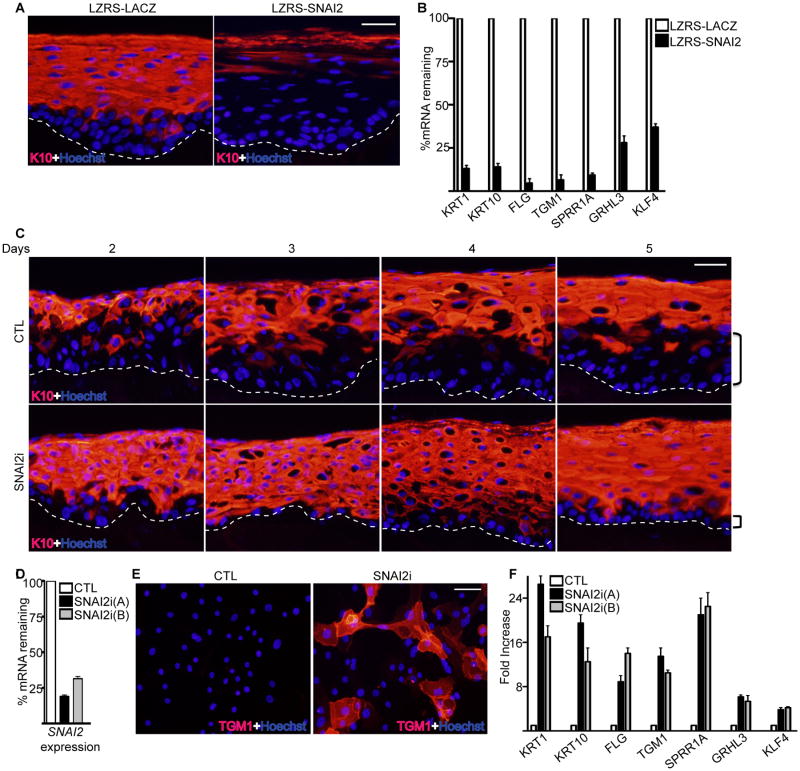
Figure 2. The levels of SNAI2 controls epidermal differentiation.
(A) Epidermal progenitor cells transduced with the LZRS retrovirus encoding either LACZ controls (LZRS-LACZ) or SNAI2 (LZRS-SNAI2) were used to regenerate human epidermis by placing the cells on devitalized human dermis. Keratin 10 (K10) staining shown in red marks the differentiated epidermal layers. Hoechst staining in blue marks the nuclei. The dashed lines denote basement membrane zone (Scale bar=40μm; n=3 regenerated human epidermis per group). (B) RT-qPCR for expression of differentiation genes from samples isolated from (A). Expression levels were normalized to GADPH. Error bars=mean with SEM. (C) Epidermal cells were transduced with retroviruses expressing shRNAs targeting either control (CTL) or SNAI2 (SNAI2i) and used to regenerate human epidermis. Tissue was harvested days 2-5 after placement of cells onto dermis to determine the kinetics of epidermal differentiation with SNAI2 loss. K10 staining is shown in red. Brackets denote size of undifferentiated basal layer. Scale bar=40μm; n=3 regenerated human epidermis per group. (D) RT-qPCR for expression of SNAI2. Cells were knocked down for SNAI2 using two distinct shRNAs. (E) Control and SNAI2i cultured primary epidermal progenitor cells were stained for differentiation protein transglutaminase I (TGM1: shown in red). Scale bar=20μm. (F) RT-qPCR for expression of differentiation genes from CTL and SNAI2i cultured cells. Knockdown of SNAI2 using two distinct shRNAs [SNAI2i (A) and SNAI2i (B)] is shown. Expression levels were normalized to GADPH. Error bars=mean with SEM.