Figure 1.
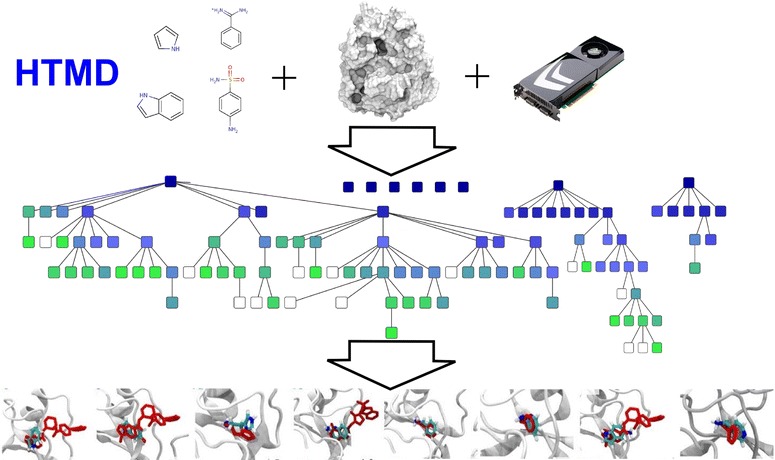
A high-throughput molecular dynamics workflow. A protein of interest is selected for study along with potential ligands (if any), and are simulated across multiple parallel runs using GPU devices (top). Additional rounds of simulation are performed, and new simulations may be respawned manually or automatically from previous runs to enhance sampling (middle). In the case of a fragment screen, for example, the result is a series of binding poses which can be compared and contrasted with other methods or used as a basis for lead development (bottom). Affinity and kinetic data are available for each interaction thanks to Markov state modelling.
