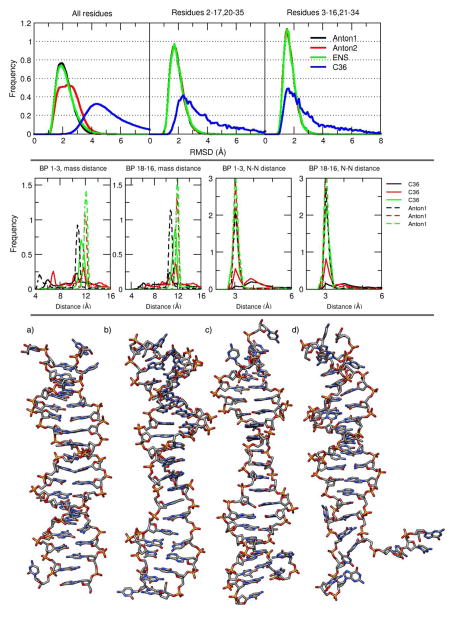
Figure 8.
Top: histogram of RMSD values for the Anton1 (black), Anton2 (red), ENS (green) and CHARMM C36 (blue). The reference for the AMBER simulation was a 10 μs average structure from Anton1. For the CHARMM simulation, the reference used was also a 10 μs average structure obtained from the full CHARMM simulation. Middle: normalized histograms of the distance vs. time of end base pairs for CHARMM and Anton1. Each of the plots represents a histogram of the distance vs time analysis for the 3 base pairs at the end of the DNA chain, using the full CHARMM trajectory. The two plots on the middle-left were calculated using the distance between the center of mass for residues 1, 36 (black), 2, 35 (red) and 3, 34 (green) and the base pairs on the other end of the DNA chain (pairs 18,19, 17,20 and 16,21). The solid lines are from the CHARMM simulation, dashed lines are from the Anton1 simulation. The two plots on the middle-right were obtained measuring the distance between the N1 atom of guanine and N3 of cytosine which gives a good measure to determine base pair opening. Bottom: the 4 representative structures of the most populated clusters from the clustering analysis using the full C36 trajectory (no hydrogens are shown). The clustering was obtained using the average-linkage algorithm (see Table S2 for exact CPPTRAJ input).