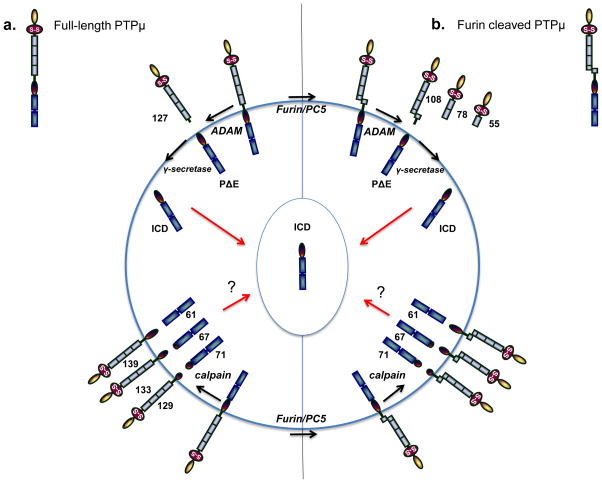
Figure 1.
PTPμ proteolysis in cancer cells as a model for R2B proteolysis. PTPμ, PTPκ, and PCP-2 have all been shown to be proteolyzed in cancer cells, and may follow the same steps as has been observed for PTPμ proteolysis. The molecular weights shown are for PTPμ. PTPμ exists at the cell surface in two full-length forms: either the non-furin processed full-length PTPμ protein or the furin-processed form that consists of two non-covalently associated PTPμ subunits (furin cleaved PTPμ). In (a), ADAM protease cuts full-length PTPμ in the extracellular domain to yield a shed 127 kDa fragment and a membrane-tethered PΔE fragment. The PΔE fragment is then proteolyzed by the γ-secretase complex to release the ICD fragment from the plasma membrane. The ICD is capable of translocating into the cell nucleus. PTPμ is proteolyzed differently in cancer cells in response to ionomycin stimulation. Full-length PTPμ is also cleaved by calpain inside of the cell, to yield three membrane-free fragments of 71, 67, and 61 kDa and three integral membrane forms of PTPμ with intact extracellular domains of 129, 133, and 139 kDa. In (b), furin cleaves PTPμ in the golgi network to yield two non-covalently associated subunits at the plasma membrane. Furin-processed forms of PTPμ are also cleaved in the extracellular domain by ADAM to yield shed fragments of 108 kDa. The membrane-tethered fragment is then cleaved by the γ-secretase complex to yield the ICD fragment. In cancer cells, calpain cleavage of furin processed PTPμ yields membrane-free cytoplasmic fragments of 71, 67, and 61 kDa, as is observed for full-length PTPμ (a). Three integral membrane forms of PTPμ with intact extracellular domains are also generated in response to calpain cleavage. PTPκ is proteolyzed by a furin-like protease, an ADAM, and the γ-secretase complex, just like PTPμ.