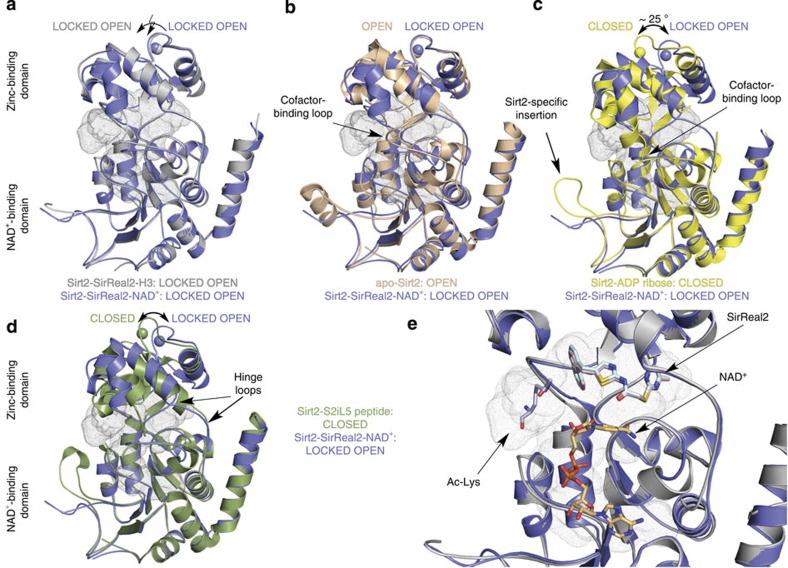
Figure 2. SirReal2 functions as a molecular wedge locking Sirt2 in an open conformation.
(a) Overlay of Sirt2–SirReal2–NAD+ (slate blue) and Sirt2–SirReal2–H3 (light grey). Both structures are very similar (r.m.s.d. (Cα atoms)=0.8 Å) and feature an open conformation. The active site is indicated by small grey dots. (b) Superposition of Sirt2–SirReal2–NAD+ (slate blue) with Sirt2–apo (PDB-ID 3ZGO, salmon, residues 34–45 are omitted for better clarity). Both structures feature an open state despite major structural differences in the zinc-binding domain. (c) Superposition of Sirt2–SirReal2–NAD+ (slate blue) with the Sirt2-ADPR complex (PDB-ID 3ZGV, yellow, residues Tyr139-Gly141 of one hinge loop were not defined in the electron density map). The structures display major conformational differences in the orientation of the zinc-binding domain. While the ADPR complex is in a closed state, Sirt2–SirReal2–NAD+ adopts an open state. (d) Superposition of Sirt2–SirReal2–NAD+ with Sirt2 in complex with a macrocyclic peptide inhibitor S2iL5 (PDB-ID 4L3O, green). Similar to the Sirt2–ADPR complex, the Sirt2–S2iL5 complex assumes a closed conformation. While the Rossmann fold domain is very similar in both structures, major structural differences can be seen at the zinc-binding domain and at the Sirt2-specific insertion. (e) Close-up view on the active site using the superposition shown in a. SirReal2 (Sirt2–SirReal2–NAD+, light pink sticks; Sirt2–SirReal–H3, light cyan sticks) occupies the extended C-site of Sirt2. Binding of SirReal2 neither prevents binding of the acetyl-lysine substrate (light blue sticks) nor the cosubstrate NAD+ (light orange sticks). The cofactor-binding loop of both structures is omitted for clarity.