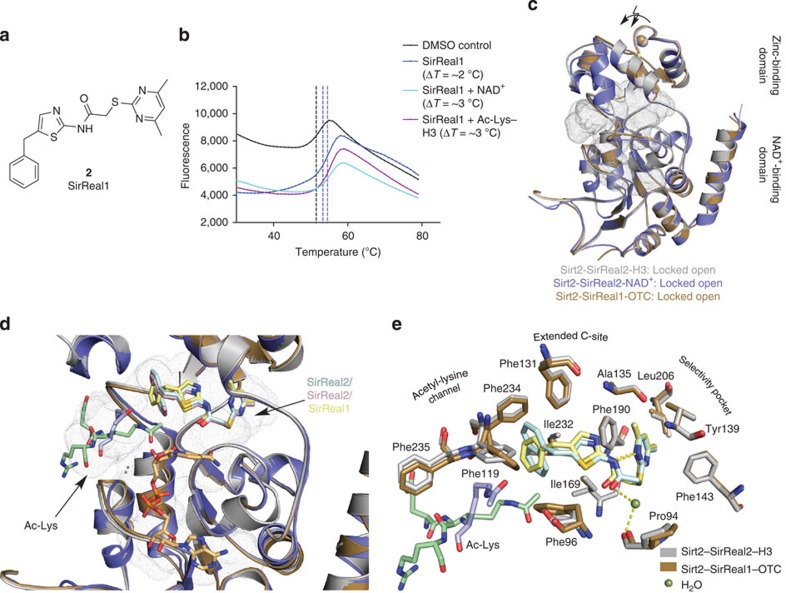
Figure 4. SirReal1 selectively inhibits Sirt2 and functions as a molecular wedge to lock Sirt2 in an open conformation.
(a) Chemical structure of SirReal1 (2). (b) Representative thermal stability plots for Sirt2 in the presence of SirReal1 (50 μM) and either the cofactor NAD+ (5 mM) or an acetyl-lysine H3 peptide (5 mM). The presence of the cosubstrates enhances the stabilization of the Sirt2–SirReal1 complex (n=3). Representative thermal stability plots of Sirt2 in the absence of SirReal2 and the presence of NAD+ or an acetyl-lysine oligopeptide are shown in Supplementary Fig. 1d. (c) Overlay of Sirt2–SirReal1–OTC (brown) with Sirt2 structures in complex with SirReal2 (Sirt2–SirReal2–H3, light grey; Sirt2–SirReal2–NAD+, slate blue). All Sirt2–SirReal complexes share a high similarity (r.m.s.d. (Cα atoms)=0.44 Å to Sirt2–SirReal2–H3, 0.59 Å to Sirt2–SirReal2–NAD+) and represent the locked open conformation. The active site is represented as grey dots. (d,e) SirReal1 (light yellow sticks) occupies the extended C-site in a very similar fashion as observed for SirReal2 (light blue in Sirt2–SirReal2–H3, light pink in Sirt2–SirReal2–NAD+). Differences can be observed for the position of the side chains of Phe119, Phe235 and the acetyl-lysine peptides. The acetyl-lysine-binding site as well as the selectivity pocket are also the sites of major conformational changes compared with Sirt2–apo (PBD-ID 3ZGO) and Sirt2-ADPR (PDB-ID 3ZGV, see Fig. 3e). Hydrogen bonds are shown in dashed yellow lines. The cofactor-binding loop of d is omitted for clarity. A stereo image of the σ-weighted 2Fo−Fc electron density maps for SirReal1 and the Ac-Lys-OTC oligopeptide as well as σ-weighted Fo−Fc electron density OMIT maps of both ligands are shown in Supplementary Fig. 5b,d.